Figure 7.
HRIP1 Gene Deletion in A. alternata R2 and Virulence Phenotype of Mutants Compared with R2 on Wild-Type ‘Greensleeves’ Apple Leaves.
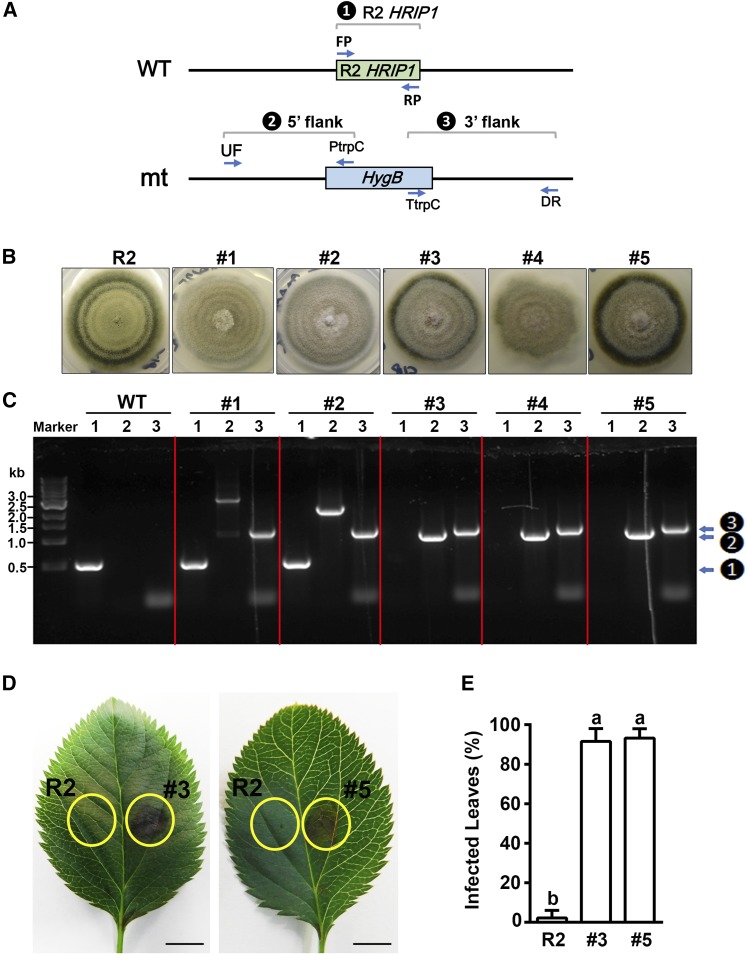
(A) Scheme of HRIP1 gene deletion from wild-type A. alternata R2 (WT) and three diagnostic PCR reactions (1 to 3, inside black circles, here, and in [C], right side) to identify deletion mutants (mt).
(B) Morphological phenotype of wild-type strain R2 and five HygBR candidate transformants (#1 to #5).
(C) Diagnostic PCR analysis using DNA of wild-type R2 and the five transformants as template. Lanes 1 to 3 in each panel show reactions with primer pairs FP/RP (detects HRIP1), UF/PtrpC (detects targeted insertion into the 5′ flank of HRIP1), and TtrpC/DR (detects insertion into the 3′ flank of HRIP1), respectively. Marker is a 1-kb DNA ladder. Note candidates #1 and #2 still have HRIP1 bands, while #3 to #5 do not (1 to 3, inside black circles, right side).
(D) Leaf phenotypes after R2 and mutant #3 or #5 inoculations, with the injection site at the center of each yellow circle. Bars = 5 mm.
(E) Percentages of infected leaves after inoculation. Data are mean ± sd of three biological replicates with 40 leaves pooled from 20 in vitro shoots per replicate. Different letters (a and b) indicate significant differences (P < 0.01) using Duncan’s MRT after ANOVA.