Figure 1.
KCH Is Required for Protonema Growth, Gametophore Leaf Morphology, and Rhizoid Elongation.
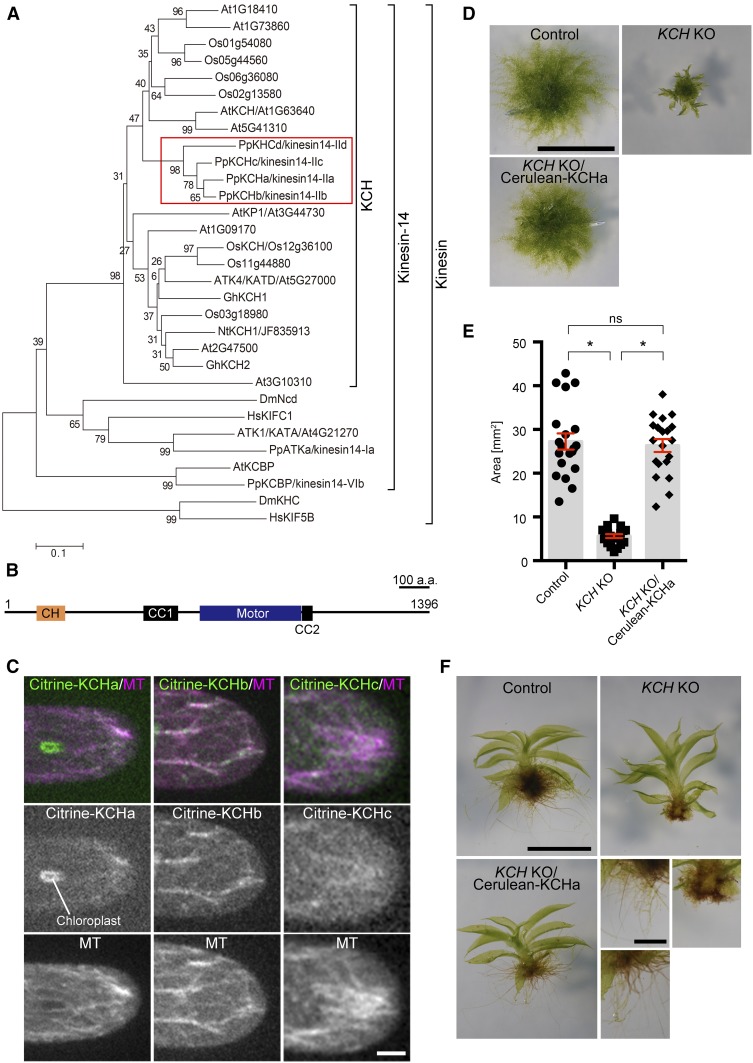
(A) Phylogenetic tree of plant KCH subgroup members and other members of the kinesin family. At, Arabidopsis thaliana; Os, Oryza sativa; Pp, Physcomitrella patens; Gh, Gossypium hirsutum; Nt, Nicotiana tabacum; Dm, Drosophila melanogaster; Hs, Homo sapiens. P. patens genes are highlighted with a red box. Horizontal branch length is proportional to the estimated evolutionary distance. Bar = 0.1 amino acid substitutions per site.
(B) Schematic diagram of domain organization and coiled-coil (CC) prediction of P. patens KCHa (1–1396 amino acids). CH domain (106–199 amino acids) and kinesin motor domain (642–975 amino acids) were predicted by an NCBI domain search. Predicted coiled-coil domains with a probability >0.7 and 21 residue windows (Coiled-Coils Prediction; PRABI Lyon Gerland) are displayed (454–567 amino acids and 978–1012 amino acids).
(C) MT localization of Citrine-KCHa, -KCHb, and -KCHc at the caulonemal cell tip. Image contrast was individually adjusted for each sample. Bar = 5 µm.
(D) and (E) Colony size comparison between control (parental line expressing GFP-tubulin/histoneH2B-mRFP), KCHabcd KO (KCH KO), and KCH KO/Cerulean-KCHa. Colonies were cultured for 3 weeks from the stage of protoplasts. Bar in (D) = 5 mm. Bars and error bars in (E) represent the mean and se, respectively. Control, n = 20; KCH KO, n = 20; KCH KO/Cerulean-KCHa, n = 20. *P < 0.0001; ns (not significant), P > 0.7 (unpaired t test with equal sd, two-tailed). Experiments were performed three times and the data analyzed twice. The data of one experiment are displayed.
(F) Gametophores and rhizoids cultured for 4 weeks on BCDAT medium. Bars = 1 mm (top) and 0.5 mm (bottom).