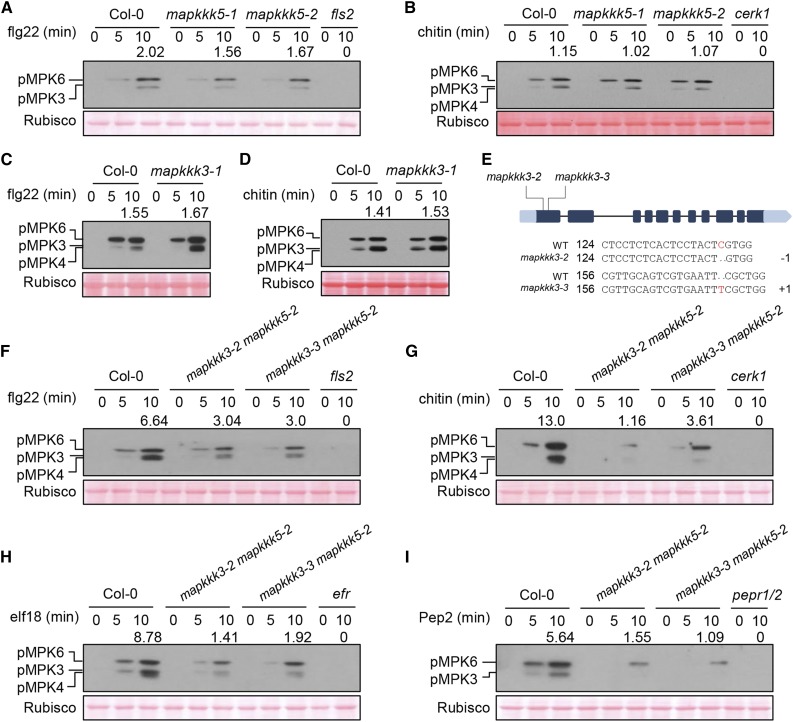
Figure 2.
MAPKKK3 and MAPKKK5 Regulate Pattern-Induced MAPK Activation.
(A) to (D) Pattern-triggered MAPK activation is normal in mapkkk5 ([A] and [B]) and mapkkk3 ([C] and [D]) single mutants. Ten-day-old seedlings were sprayed with flg22 or chitin, and samples were harvested at the indicated times for immunoblot analysis with anti-pERK antibody. Note that MPK4 and MPK3 are highly similar in size and were not well-separated in some experiments. Numbers indicate arbitrary densitometry units of phosphorylated MPK3/6 (pMPK3/6) bands normalized to Rubisco 10 min after pattern stimulation. All assays were performed three times, and a representative photograph is shown.
(E) CRISPR-Cas9-mediated mutations in the first exon of MAPKKK3 in the mapkkk3 mapkkk5 (mapkkk3-2 mapkkk5-2 and mapkkk3-3 mapkkk5-2) double mutant lines. The numbers 124 and 156 indicate the nucleotide positions in the MAPKKK3 coding sequence. −1 and +1 indicate frame shifts in the mutant lines.
(F) to (I) Pattern-triggered MPK3/6 activation is diminished in mapkkk3 mapkkk5 double mutants. Seedlings were sprayed with flg22 (F), chitin (G), elf18 (H), or Pep2 (I), and immunoblot analysis with anti-pERK antibody was performed at the indicated times. Note that MPK4 and MPK3 are highly similar in size and were not well separated in some experiments. Numbers indicate relative protein band density of pMPK3/6 normalized to the loading control (Rubisco). All assays were performed at least three times, and a representative photograph is shown. Ponceau staining of Rubisco indicates equal loading.