Figure 2.
Potri.002G146400 Encodes an HTH Motif Containing Protein.
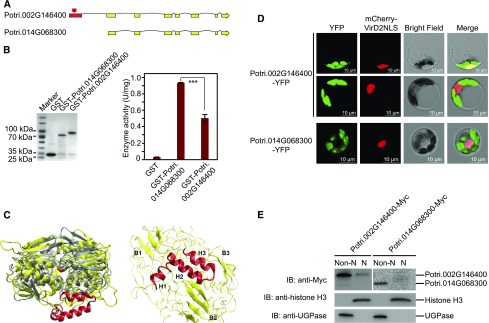
(A) Comparison of two putatively paralogous genes, Potri.002G146400 and Potri.014G068300, shows an additional exon (red) encoding an N-terminal HTH motif.
(B) Left panel: Purified GST, GST-Potri.002G146400, and GST-Potri.014G068300 were resolved on an SDS-PAGE gel and stained with Coomassie blue. The protein molecular weights are indicated on the left. GST only was used as a negative control. Right panel: EPSP synthase activities of GST, GST-Potri.002G146400, and GST-Potri.014G068300. For each sample, three reactions were performed in parallel to calculate the mean value and sd (error bar), which were used in Student’s t tests (***P < 0.001, **P < 0.01, *P < 0.05, and ns P > 0.05).
(C) Left: Potri.002G146400 (yellow) superimposed on an EPSP synthase from the Agrobacterium strain CP4 crystallographic structure (gray, PDB entry 2GG6) bound with a shikimate-3-phosphate substrate (blue and red spheres). The HTH domain is shown in red. Right: Detailed view of the HTH domain of Potri.002G146400 comprised of three characteristic α-helices H1 to H3, which are surrounded by β-sheets B1 to B3 in the enzyme. The rest of the enzyme is shown in ribbons.
(D) Subcellular localization of Potri.002G146400-YFP and Potri.014G068300-YFP in Populus protoplasts (green). The nuclear marker mCherry-VirD2NLS is shown in red. Bar = 10 µm.
(E) Immunoblot showing the accumulation of Potri.002G146400-Myc in non-nuclear and nuclear fractions. Potril002G146400-Myc and Potri.014G068300-Myc were blotted using anti-Myc. UGPase and histone H3 were blotted to indicate the purity of non-nuclear and nuclear fractions, respectively. Non-N, non-nuclear fraction; N, nuclear fraction. P value comparison is calculated using two-tailed Student’s t tests (***P < 0.001, **P < 0.01, *P < 0.05, and ns P > 0.05).