Figure 2.
Loss of 24-Nucleotide siRNAs and Gain of 21- and 22-Nucleotide siRNAs in Double Mutants.
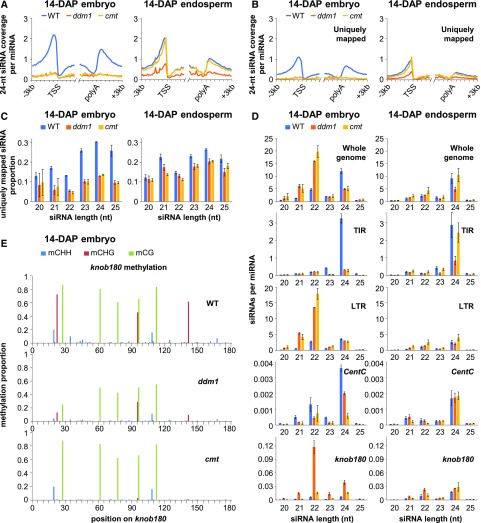
(A) The 24-nucleotide siRNA coverage near genes. All genes were defined by their annotated transcription start sites (TSS) and polyadenylation sites (polyA) and split into nonoverlapping 100-bp intervals. The siRNA coverage for each 100-bp interval was summed for the complete set of genes and normalized by miRNAs.
(B) Uniquely mapped 24-nucleotide siRNA coverage near genes, as in (A).
(C) Proportion of uniquely mapped siRNAs for each length.
(D) Length distribution of various classes of siRNAs. “Whole genome” includes all mapped siRNAs. “TIR” includes all siRNAs that overlapped by at least half their lengths to Mutator, hAT, Harbinger, or Mariner TIR transposons. “LTR” includes all siRNAs that overlapped by at least half their lengths with Gypsy or Copia LTR retrotransposons. CentC includes all siRNAs that aligned to a CentC consensus sequence and knob180 includes all siRNAs that aligned to a knob180 consensus sequence. siRNA counts were normalized per miRNA. Error bars are standard errors of the means for the biological replicates of each genotype.
(E) Single-base-pair DNA methylation in knob180 repeats. WGBS reads were mapped to the knob180 consensus sequence and methylation calculated as the proportion of methylated C over total C in each sequence context.