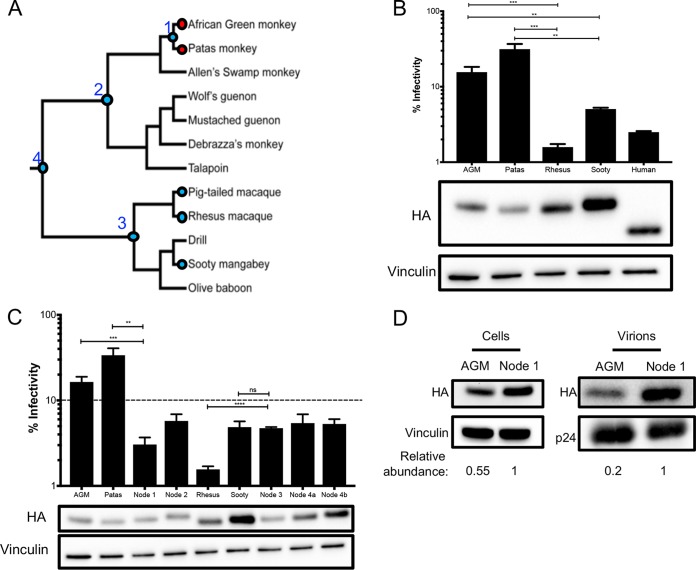
FIG 4.
AGM ancestors produce potent antiviral proteins. (A) Phylogeny, depicted as a cladogram, based on the accepted species tree of all sequenced Old World primates included in the study (28). The blue circles denote active antiviral proteins; the red circles denote less active antiviral proteins. Ancestral nodes are labeled with numbers (1 to 4). (B) (Top) Single-cycle infectivity assay for HIVΔvif against extant primate A3H proteins. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. Statistical differences were determined by unpaired t tests. (Bottom) Western blot analysis of the protein expression level. Vinculin was used as a loading control. Human A3H is missing 28 C-terminal amino acids due to a natural premature stop codon (15) and thus ran lower on the blot. (C) (Top) Single-cycle infectivity assay for HIVΔvif against ancestral A3H proteins and their extant descendants. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. The dashed line at 10% is an arbitrary reference point. (Bottom) Western blot analysis of the protein expression level. Vinculin was used as a loading control. (D) Packaging of A3H proteins into virions analyzed by Western blotting. Relative abundance in cellular expression (left) and virion incorporation (right) were determined compared to the node 1 A3H ancestor. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; ns, not significant.