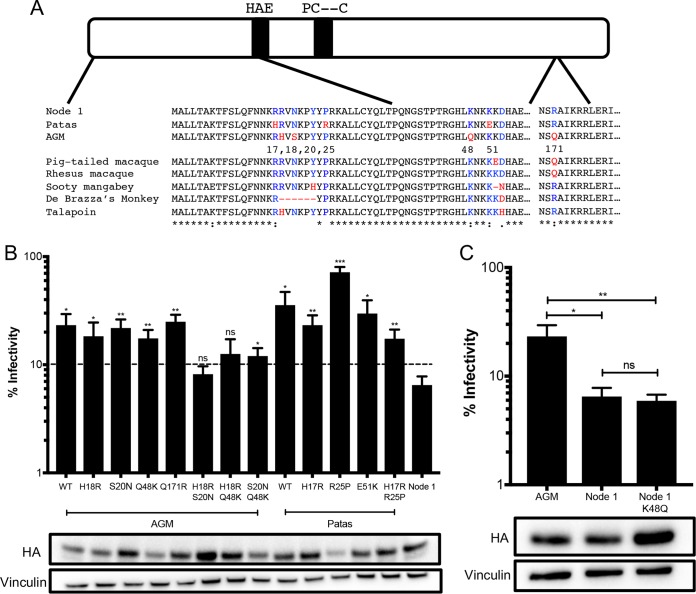
FIG 5.
Multiple amino acid mutations required for an increase in antiviral activity. (A) Schematic of the A3H protein. The black bars outline the A3H catalytic site. Numbered amino acid residues that are different between AGM A3H, patas A3H, and the node 1 ancestor are in red in the protein sequence alignment, and ancestral residues are in blue. Amino acids that are different in other primates are similarly colored. (B) (Top) Single-cycle infectivity assay for HIVΔvif against extant mutants. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. Statistical differences were determined by unpaired t tests: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant. Statistically significant differences from the node 1 ancestor are depicted. (Bottom) Western blot analysis of protein expression levels of HA-tagged extant mutants made in the AGM and patas A3H backgrounds. Vinculin was used as a loading control. (C) (Top) Single-cycle infectivity assay for HIVΔvif against AGM haplotype 1, node 1 ancestor, and node 1 mutant A3H. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. Statistical differences were determined by unpaired t tests: *, P ≤ 0.05; **, P ≤ 0.01; ns, not significant. (Bottom) Western blot analysis of protein expression levels of HA-tagged proteins made in the AGM and patas A3H backgrounds. Vinculin was used as a loading control.