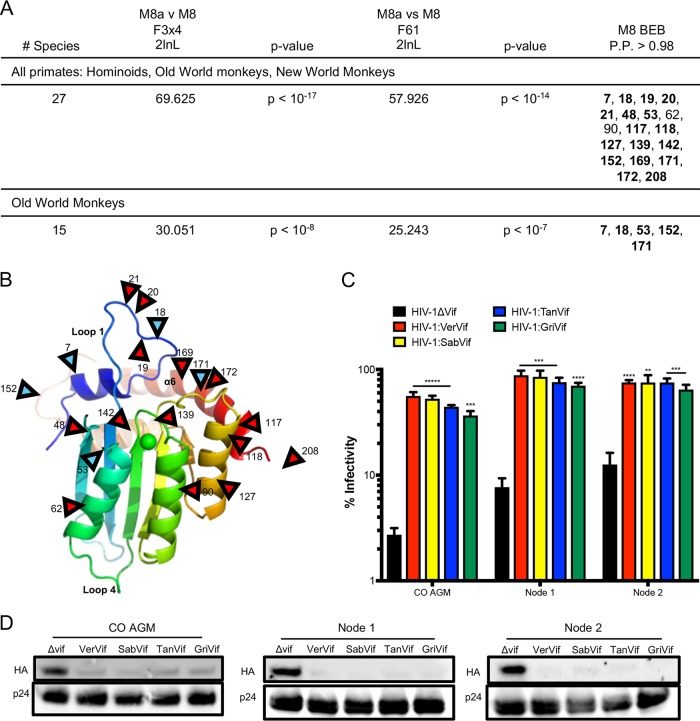
FIG 6.
Evolution of A3H is not driven by Vif. (A) Results of positive-selection analyses of primate A3H. The far-right column lists sites under positive selection with dN/dS values of >1 with a posterior probability of >0.98 under M8 Bayes empirical Bayes (BEB) implemented in PAML model 8. The sites are relative to African green monkey A3H. Sites that had a posterior probability of >0.98 in both codon models (F3x4 and F61) are in boldface. (B) A3H from individual PR01190 was modeled onto the pig-tailed macaque A3H structure (Protein Data Bank [PDB] accession no. 5W3V). Locations of positively selected sites are indicated by red arrowheads. The blue arrowheads indicate sites also under positive selection in Old World primates. Site 208 is part of a region missing from the crystallized protein and is therefore not resolved in the model. (C) Single-cycle infectivity assay done with HIVΔvif and HIV-1 expressing SIVagm Vif in the presence of codon-optimized AGM A3H haplotype 1 (CO AGM), the node 1 ancestor, and the node 2 ancestor. Relative infection was normalized to viral infectivity in the absence of A3 proteins. Averages of three replicates, each with triplicate infections (+SEM), are shown. Statistical differences were determined by unpaired t tests: **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. Statistically significant differences from the restriction of HIVΔvif are depicted. (D) Western blot analyses demonstrating virion incorporation of A3H proteins in the absence and presence of Vif. p24 was used as a loading control.