FIG 6.
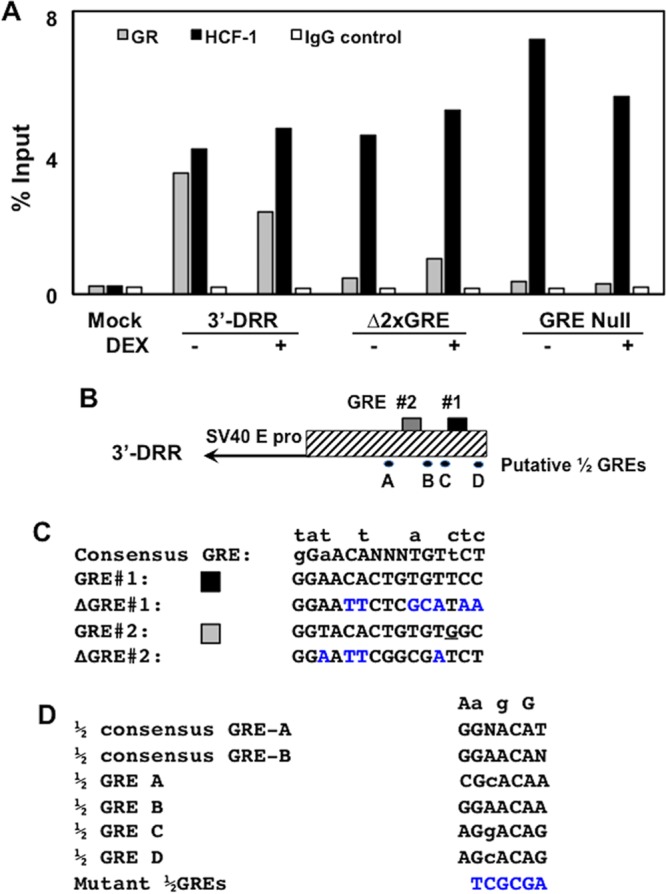
HCF-1, but not GR, occupies sequences in 3′-DRR when GRE sites are mutated. (A) Neuro-2A cells were cotransfected with the plasmid expressing the mouse GR and the 3′-DRR construct. Cultures were subsequently treated with 10 μM DEX (+) for 3 h at 45 h after transfection. HCF-1 and GR occupancy were assessed by ChIP assays using the DRR primer set. IgG, control; input, 10% of total DNA; M, DNA size markers. The intensity of bands was quantified using a Bio-Rad ChemiDoc system. The trends in this ChIP assay are consistent with results from three independent studies. (B) Schematic of the 3′-DRR promoter construct and location of the two GREs and four ½ GREs. (C) Sequence of the consensus GRE, the two GREs in the IEtu1 promoter, and mutants. Nucleotides above the consensus GRE denote changes that still result in GR binding and transactivation. Blue nucleotides denote mutations introduced into GRE#1 and GRE#2 (26). The underlined G in GRE #2 is a mismatch to the consensus GRE. (D) The two consensus ½ GREs (GRE-A and GRE-B) were previously described (35), and nucleotides listed above the sequences are differences observed in some ½ GREs that are capable of trans-activation by a GR monomer. Sequences of the four ½ GREs were identified in the 3′-DRR. A NruI restriction enzyme site (TCGCGA) was used to mutate each of the four ½ GRE sites (blue nucleotides).
