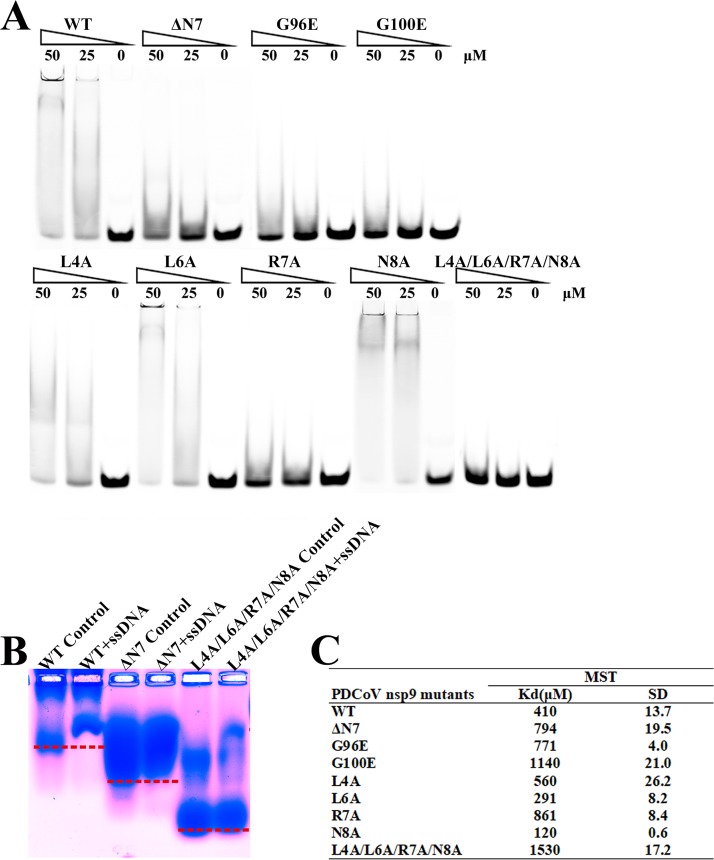
FIG 4.
The N-finger motif can affect the ssDNA binding abilities of PDCoV nsp9. (A) The ssDNA binding abilities of nsp9-WT, nsp9-ΔN7, nsp9-G96E, nsp9-G100E, nsp9-L4A, nsp9-L6A, nsp9-R7A, nsp9-N8A, and nsp9-L4A/L6A/R7A/N8A as determined by EMSA. Each protein was assayed at different concentrations (0, 25, and 50 μM) with 5 μM 5′-Cy5-labeled 36-mer ssDNA. (B) Zone-interference gel electrophoresis illustrating the association of nsp9-WT, nsp9-ΔN7, and nsp9-L4A/L6A/R7A/N8A with 36-mer ssDNA. The red dotted lines represent the position of the bands of nsp9-WT, nsp9-ΔN7 and nsp9-L4A/L6A/R7A/N8A without reaction with ssDNA. (C) The ssDNA binding affinities of nsp9-WT and mutants were measured by MST assays. Each protein was assayed in 2-fold concentration steps with 10 nM 5′-Cy5-labeled 36-mer ssDNA. The measured Kd of each protein is shown in the table.