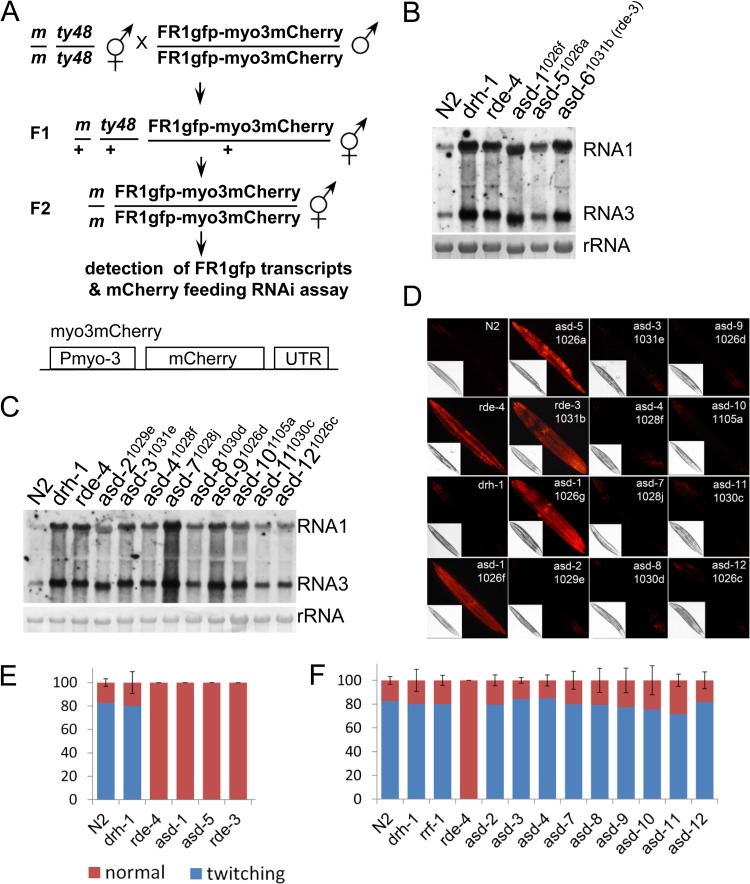
FIG 6.
Function characterization of the candidate genes. (A) Experimental design for the introduction of candidate alleles into worm strain LR11. m, loss-of-function alleles identified in the biased genetic screen. Pmyo-3, myo-3 promoter that is constitutively active in body wall muscle. (B) Northern blot detection of FR1gfp replication in LR11 worms that contain genetic alleles corresponding to asd-1, asd-5, and rde-3. The replication of FR1gfp was also detected in wild-type N2 worms and drh-1 and rde-4 mutants as references. (C) Northern blot detection of FR1gfp replication in LR11 worms that contain genetic alleles corresponding to asd-2, asd-3, asd-4, asd-7, asd-8, asd-9, asd-10, asd-11, and asd-12. (D) Visualization of mCherry fluorescence in LR11 worms that contain genetic alleles corresponding to candidate alleles, as indicated within each set of images. All worms were fed using E. coli food expressing dsRNA derived from mCherry coding sequence. The inset images were taken under white light. All images were taken using the same exposure. (E and F) unc-22 RNAi phenotype observed in LR11 worms containing genetic alleles, as indicated in panels B and C, corresponding to candidate genes. unc-22 dsRNA was injected at a concentration of 100 ng/μl. Shown here are the percentages of twitching F1 progenies of the injected worms collected between 8 and 32 h postinjection. The error bars indicate standard deviations for the twitching phenotype.