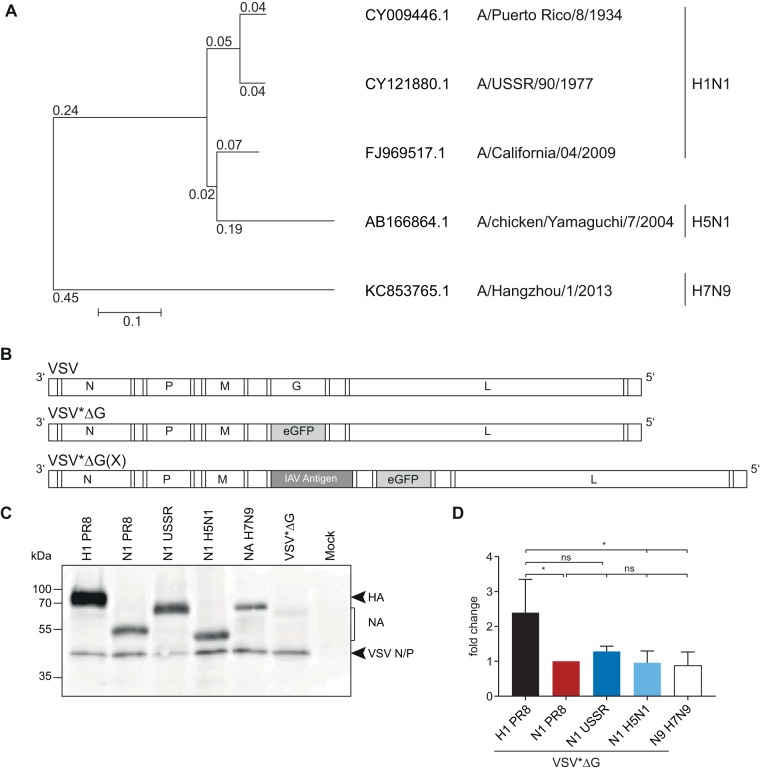
FIG 1.
Generation and characterization of influenza antigen-expressing VSV replicons. (A) Phylogenetic relationship of influenza A virus NA genes included in this study. Amino acid sequences were aligned using ClustalW and a phylogenetic tree constructed using the neighbor-joining method and 1,000 bootstrapped replications with observed amino acid differences as the distance on the aligned sequences using MEGA7. (B) Scheme of the genomes of the parental VSV and the recombinant VSV*ΔG replicons. VSV*ΔG lacks the gene encoding the VSV G protein and includes the eGFP gene in an additional transcription unit, depicted in gray. VSV*ΔG(X) encodes the respective heterologous influenza virus antigens, depicted in dark gray. (C) Vero E6 cells were surface biotin labeled with EZ-Link sulfo-NHS-biotin (Pierce) 12 h after infection with the respective VSV*ΔG replicons, followed by cell lysis and immunoprecipitation of 50 μg of cell extracts with homologous mouse antiserum. Proteins in the pellet fraction were separated on an SDS-PAGE gel, and surface biotin-labeled proteins were visualized using an avidin-HRP secondary antibody. (D) Protein bands from three independent replicates were quantified and normalized relative to NAPR8 for each blot and are shown as fold changes. Error bars represent the standard deviations of the means. Groups were compared using one-way ANOVA with a Tukey posttest, and statistical significance is indicated (*, P < 0.05; ns, not significant).