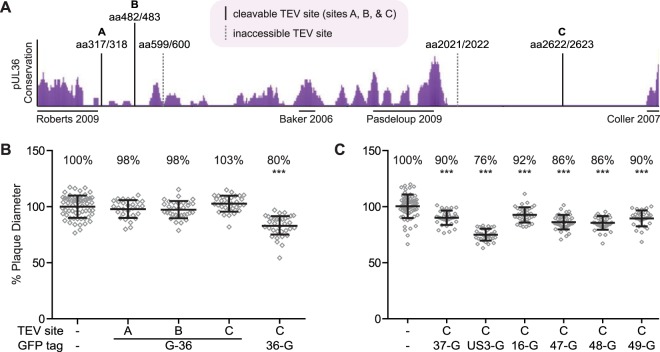
FIG 1.
Recombinant PRV strains used in this study. (A) The degree of conservation across the pUL36 predicted amino acid sequence from seven alphaherpesviruses is shown as purple peaks (eShadow). The locations of TEV site insertions are indicated by vertical lines and labeled with flanking amino acid (aa) positions. Solid lines were cleavable with exogenous TEV protease, and dotted lines were refractory to cleavage. Regions of pUL36 suggested or shown to interact with capsids are marked by horizontal lines under the conservation plot. (B and C) Plaque diameters of recombinant viruses encoding a pUL25/mCherry capsid fusion, a GFP fusion to a designated tegument protein, and a TEV site within pUL36 were compared to a parental virus containing only the pUL25/mCherry tag. Diameters are presented as a percentage of the parental virus. Error bars indicate standard deviations (***, P < 0.05 compared to wild type).