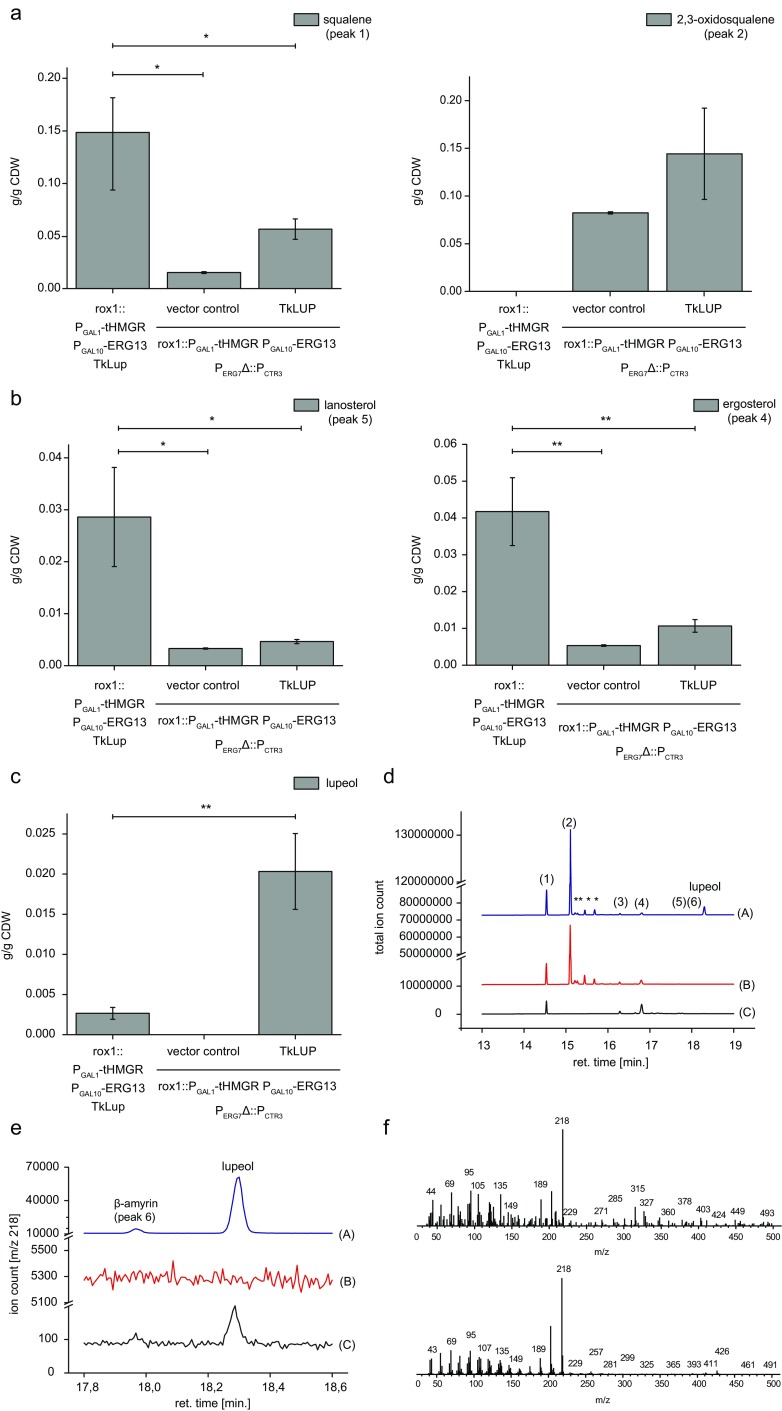
Fig. 4.
The repression of ERG7 enhances the accumulation of lupeol while reducing the abundance of sterols. a The accumulation of 2,3-oxidosqualene and the lower levels of squalene were observed in the yeast strain carrying the TkLUP sequence in addition to the rox1::PGAL1-tHMGR PGAL10-ERG13 and the PERG7Δ::PCTR3 cassettes, following exposure to 150 μM CuSO4 during growth (rox1::PGAL1-tHMGR PGAL10-ERG13 PERG7Δ::PCTR3 TkLUP) compared to the parental strain (rox1::PGAL1-tHMGR PGAL10-ERG13 TkLUP; p = 0.04422 for squalene). b The sterol content was reduced as anticipated, as shown by the amounts of lanosterol and ergosterol as representatives for sterol biosynthesis. The repression of ERG7 resulted in a 6.5-fold decrease in lanosterol levels (p = 0.02384) and a 3.9-fold decrease in ergosterol levels (p = 0.00941). c The repression of sterol biosynthesis enhances lupeol production by 7.6-fold (p = 0.00637), suggesting the redirection of the metabolic flux from sterol biosynthesis to lupeol production. d Comparison of the total ion count of different yeast strains. (A) = rox1::PGAL1-tHMGR PGAL10-ERG13 PERG7Δ::PCTR3 TkLUP; (B) = rox1::PGAL1-tHMGR PGAL10-ERG13 PERG7Δ::PCTR3 vector control; (C) = TkLUP; (1) = squalene peak; (2) = 2,3-oxidosqualene peak; (3) = cholesterol peak; (4) = ergosterol peak; (5) = lanosterol peak; (6) = β-amyrin peak; asterisk = unidentified yeast metabolites. e Detailed comparison of the β-amyrin and lupeol peak [m/z = 218] from different yeast strains. (A) = rox1::PGAL1-tHMGR PGAL10-ERG13 PERG7Δ::PCTR3 TkLUP; (B) = rox1::PGAL1-tHMGR PGAL10-ERG13 PERG7Δ::PCTR3 vector control; (C) = TkLUP. f Identification of β-amyrin in yeast following the repression of ERG7. Mass spectrum of the designated β-amyrin peak from the engineered yeast strain (rox1::PGAL1-tHMGR PGAL10-ERG13 PERG7Δ::PCTR3 TkLUP) (upper part). Mass spectrum of the measured external β-amyrin standard (lower part). Standard deviation was calculated from n = 3 individual transformants. CDW = cell dry weight; one asterisk = p ≤ 0.05; two asterisks = p ≤ 0.01