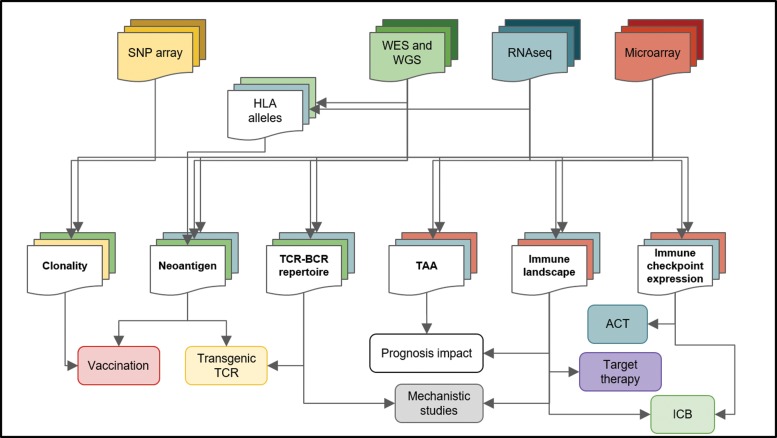
Figure 3.
Bioinformatics analysis of high-throughput data and its implications for cancer treatment. High-throughput platforms and next-generation sequencing platforms have generated large amounts of data and consists of a rich source of information for cancer research. Different techniques (top squares) are submitted to distinct pipelines, producing particular results (middle squares) that can have unique implications for cancer therapy and research (bottom squares). For instance, from microarray data (red), the immune landscape, the TAAs and immune checkpoint expression can be accessed, but neoantigens cannot be predicted. Joining multiple data sources increases the level of information about the tumor and its microenvironment. Whole-genome sequencing (WGS), whole-exome sequencing (WES), human leukocyte antigen (HLA), tumor-associated antigen (TAA), adoptive cell transfer (ACT), immune checkpoint blockade (ICB), T cell receptor (TCR), B cell receptor (BCR).