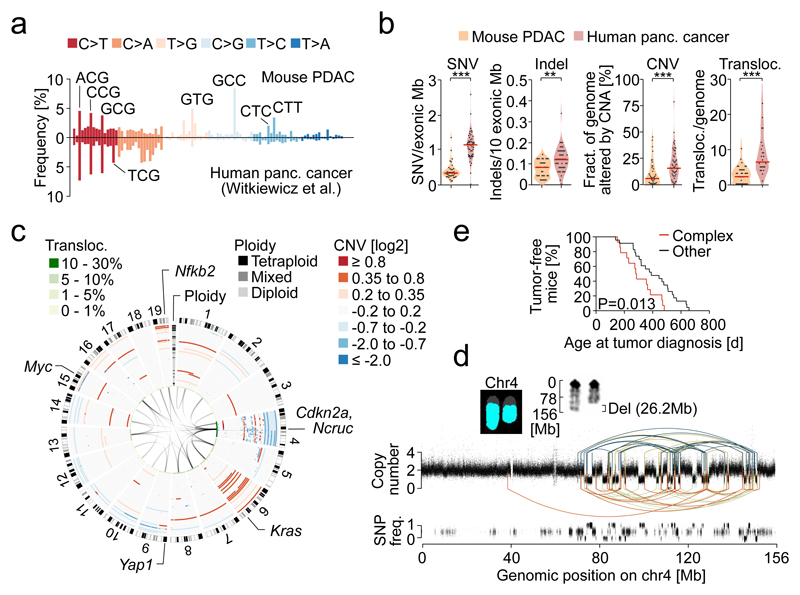
Figure 1. Genetic landscape of mouse PDAC and comparison to the human disease.
a, Trinucleotide context-dependent SNV frequencies in mouse (n=38 PK mice) and human PDAC (n=51 patients from6) derived from WES. b, SNV, indel, CNA and translocation burdens by WES, aCGH and M-FISH in PK mice (n=38) and human PDAC (n=51 patients for SNV, indel, CNA [data from6] and n=24 cell lines for translocations). **P=0.002, ***P≤0.001, two-sided Mann-Whitney test; bars, median. c, CNAs, ploidy and translocations in PK mice (n=38), detected by aCGH and M-FISH. Mixed ploidy, n≥3 diploid/tetraploid cells in 10 karyotypes. d, Rearrangement graph showing chr4 chromothripsis in mPDAC S821, based on WGS. Haplotype-specific chromosome content loss confirmed by M-FISH (n=10/10 karyotypes). e, Age at tumor diagnosis of mice having cancers with (n=14) or without (n=23) complex/clustered chromosomal rearrangements (n≥10 CNAs/chromosome). Two-sided log-rank test.