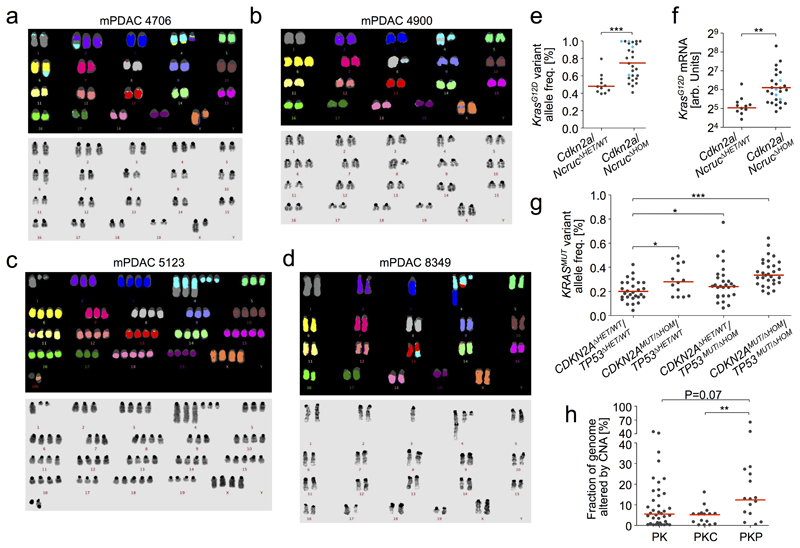
Extended Data Figure 5. Characterization of Cdkn2a (chr4) alterations and correlation with KrasMUT gene dosage variation and mRNA expression in mouse and human PDAC.
a-d, Cdkn2a alteration on mouse chr4 can occur through arm-level, complex or focal loss as well as uniparental disomy (see Figure 3). In addition, chr4 is frequently involved in inter-chromosomal translocations. Examples of representative karyotypes of primary pancreatic cancer cultures derived from PK mice with translocations involving chr4, likely affecting the Cdkn2a locus. In all 4 cases, chr4 translocations were found in all 10 metaphase spreads of each cancer, indicating their early acquisition during tumor evolution. a, mPDAC 4706 with diploid karyotype: 42, XX, del(X), +2, der(2)t(2;4)is(2;4), der(4)t(4;6)*2, +der(4)t(2;4), der(6)t(4;6). b, mPDAC 4900 also features a diploid karyotype: 41, XX, der(X)is(X;4), der(4)is(4;8), del(4), +6, der(8)t(4;8). c, mPDAC 5123 underwent polyploidization, after translocation of chr4 with chr1 and an deletion on the other copy: 78, XXXX, -1, del(1)*2, -2, +4*2, der(4)t(1;4)*3, del(4)*3, -5, -7, -9, +15, -17, +18 d, mPDAC 8349 shows a diploid karyotype: 40,XX, der(4)t(3;4), der(4)t(4;13), +del(4), der(13)t(4;13). e, KrasG12D variant allele frequencies detected by amplicon-based deep sequencing of the Kras locus are higher in Cdkn2a/NcrucΔHOM mPDAC (n=27) as compared to Cdkn2a/NcrucΔHET/WT (n=11) pancreatic cancers. All cancers are from PK mice. Blue dots indicate tumors with complete Ncruc deletion. ***P≤0.001, two-tailed Mann-Whitney test; bars, median. f, Allele-specific expression of mutant KrasG12D mRNA is increased in primary tumors from PK mice with Cdkn2a/NcrucΔHOM (n=27) background in comparison to Cdkn2a/NcrucΔHET/WT (n=11) cancers. Primary mPDACs with homozygous loss of Ncruc are highlighted in blue. KrasG12D expression was analyzed by combining amplicon-based RNA-Seq and qRT-PCR (as described in the Methods section). **P=0.003, two-tailed Mann-Whitney test; bars, median. g, KRASMUT variant allele frequencies based on WES in a published dataset of microdissected human PDAC (Witkiewicz et al., reduced stromal content) was analyzed with respect to CDKN2A and TP53 status. KRASMUT allele frequency was higher in mutated/homozygous deleted CDKN2A and/or TP53 (CDKN2AMUT/ΔHOM/TP53MUT/ΔHOM; hPDACs as compared to cancers with CDKN2AΔHET/WT/TP53ΔHET/WT status (from left: n=28, n=14, n=28, n=30). Two-sided rank-based ANOVA (P=5.8*10-6); post hoc testing with two-sided Tukey honest significant difference test, *adj. P≤0.05, ***adj. P≤0.001; bars, median. h, Fraction of the genome altered by copy number changes detected by aCGH in primary mPDACs of PK (n=38), PKC (n=16) and PKP (n=16) mice. PKP mice show a significantly increased CNA load as compared to PKC mice. Two-sided rank-based ANOVA (P=0.01); post hoc testing with two-sided Tukey honest significant difference test, **adj. P=0.009, adj. P-values for group wise comparisons are shown; bars, median. Del, deletion; der, derivative chromosome; is, insertion; t, translocation; „-“, chromosome loss; „+“, chromosome gain.