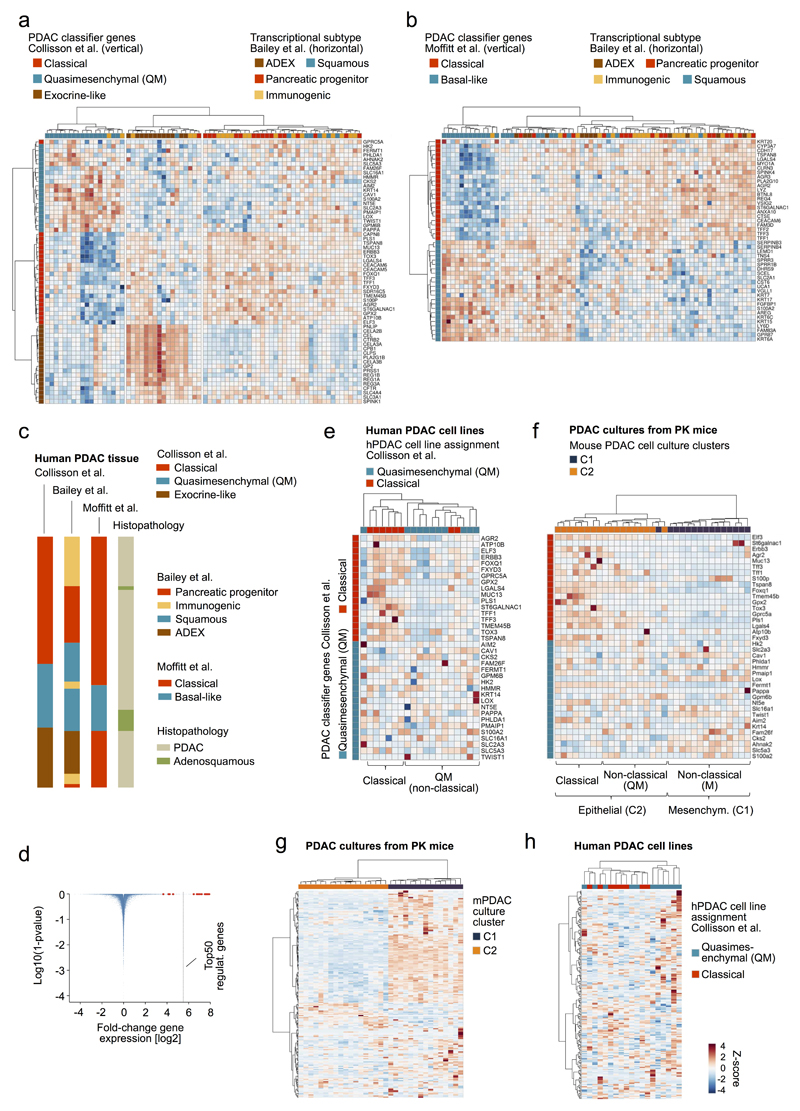
Extended Data Figure 7. Transcriptome-based subtyping of human primary pancreatic cancer and classification of human PDAC cell lines and primary PDAC cell cultures from PK mice.
a-c, Independent cross-comparison of transcriptional classification systems from Collisson et al.28, Moffitt et al.29 and Bailey et al.7. Collisson et al. performed PDAC microdissection and defined 3 transcriptional subtypes: classical, quasimesenchymal (QM) and exocrine-like. Moffitt et al. defined 2 subtypes (classical, basal-like) using (i) virtual separation of tumor and non-tumor gene expression patterns, (ii) transplantation studies and (iii) human PDAC cell lines; and proposed that the exocrine-like signature stems from exocrine pancreatic cells, rather than from the cancer cells. Bailey et al. used bulk tumors and defined 4 subtypes (pancreatic progenitor, immunogenic, squamous, aberrantly differentiated endocrine exocrine [ADEX]). RNA-Seq data from PDAC and adenosquamous pancreatic carcinoma from Bailey et al. was used for cross-comparison of classification systems. Other histological subentities of pancreatic cancer were excluded (e.g. IPMN, MCN, acinar cell carcinoma). The Bailey subtyping for this dataset was available. a, Unbiased hierarchical clustering of primary pancreatic cancer samples (n=71) from Bailey et al. using Collisson classifier genes. b, Subtyping of primary pancreatic cancer samples (n=71) from Bailey et al. using classifier genes defined by Moffitt et al. c, Consensus clustering based on analyses performed in a/b. There is considerable overlap between at least two subtypes, which are in large parts captured by the initially proposed Collisson classical and quasimesenchymal (QM) signatures (which are also detected in mouse and human PDAC cell lines; see Extended Data Figure 7e-h). The Bailey classification (based on bulk tissue analyses) suggests that Collisson classical cancers (microdissected cancer tissue) can be further sub-stratified in some with and some without a strong immune cell infiltration. The Moffitt classification suggests that the Collisson exocrine-like signature (Bailey ADEX subtype) stems from “contaminating” healthy exocrine pancreatic cells, based on the evidence described above. Given that the Collisson exocrine-like signature was derived from microdissected PDAC, such “contamination” is only conceivable, if exocrine-like signature genes were dramatically higher expressed in pancreatic acinar cells as compared to PDAC cells. d, Volcano plot showing strongly upregulated expression of exocrine-like genes in human wild-type pancreas (13 to 241 fold; median: 183-fold upregulation). Note that 15 out of 19 exocrine-like signature genes (red dots) are among the top50 genes upregulated in human wild-type pancreas (n=3) as compared to hPDAC cell lines (n=30) (y axis is calculated on Benjamini-Hochberg adj. P-values derived from R package limma [see Methods section]). Although these data do not exclude the existence of exocrine-like PDACs, they support the possibility that “contamination” with few acinar cells can impose an exocrine-like signature on a cancer. This might explain why human or mouse PDAC cell lines don´t cluster into the exocrine-like subtype (see also Extended Data Figure 7e-f below). e, Hierarchical clustering of microarray-based expression profiles using Collisson identifier genes28 on human PDAC cell lines (n=19, GEO series GSE17891). As also described earlier by Collisson et al., only two subtypes can be detected in human cell line collections: classical and quasimesenchymal (QM). Of note, the most prominent change in the QM cell lines is downregulation (extinction) of the classical assigner genes, whereas expression of QM classifier genes is quite variable. We therefore also use here the terms classical and non-classical. f, Projection of the Collisson classifiers on mouse PDAC cell culture transcriptomes (n=33) also identified classical and non-classical subtypes. The non-classical subtype contained a subset of mPDAC cell cultures from cluster C2a/b/c (epithelial morphology; equivalent of human QM) and all cluster C1 mPDACs (mesenchymal morphology; “M” cluster). g, Application of a human EMT hallmark gene set52 for hierarchical clustering of expression profiles from primary PDAC cultures (PK mice; n=33) resulted in a separation of C1 (mesenchymal) and C2a/b/c (epithelial) cell lines. h, Projection of the EMT hallmark gene set on human PDAC cell line transcriptomes (n=19, GEO series GSE17891) did not result in a clear separation of samples, indicating underrepresentation of the mesenchymal M subtype (equivalent to murine C1/“M”) in available human cell line collections. As shown in Extended Data Figure 9b, however, the EMT signature is detectable in undifferentiated human pancreatic carcinoma, which is the human equivalent of the mesenchymal mouse PDACs in C1.