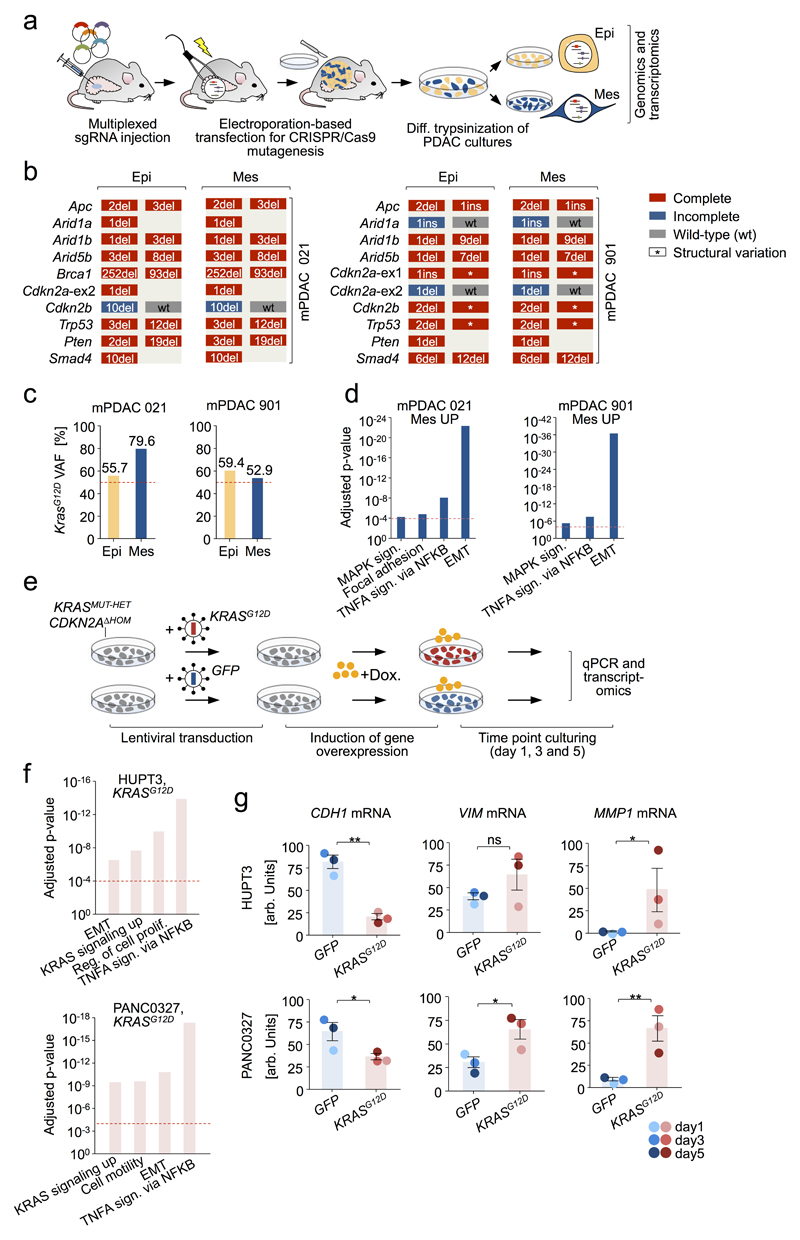
Extended Data Figure 8. Functional analyses to study the role of KrasG12D gene dosage increase in EMT. a-d, Multiplexed somatic CRISPR/Cas9 mutagenesis for phylogenetic tracking of epithelial/mesenchymal mPDAC clones in vivo.
a, Graphic demonstrates major steps of multiplexed gene editing by pooled delivery of CRISPR/Cas9 vectors, each targeting a different tumor suppressor gene in the pancreas of PK mice. Electroporation-based transfection enables low-frequency mosaic vector delivery (average of 120 cells per pancreas are transfected) to induce clonal tumors. Primary tumor cell cultures were screened for the simultaneous presence of epithelial and mesenchymal cells. Two such cancers were identified (mPDACs from mouse 021 and mouse 901) and subjected to differential trypsinization in order to enrich for each morphology. b, Amplicon-based deep sequencing of all sgRNA-targeted loci revealed identical indel patterns in both epithelial/mesenchymal culture pairs. This shows (i) that epithelial and mesenchymal cells originate from the same clone and (ii) that the CRISPR-induced mutations are not contributing to the differential phenotype. c, KrasG12D variant allele frequencies in epithelial and mesenchymal cell cultures from mPDAC 021 and mPDAC 901, as detected by amplicon-based deep sequencing. Both cancers had increased KrasG12D expression in mesenchymal cells (see Fig. 5e). In mPDAC 021 this is due to selective amplification of the KrasG12D allele in mesenchymal cells. In mPDAC 901 genetic KrasG12D amplification was not observed, suggesting induction of increased Kras expression in mesenchymal cells by other mechanisms. d, Gene set enrichment analysis using “Molecular Signatures Database” (MSigDB) of differentially regulated genes in mesenchymal versus epithelial mPDACs based on RNA-Seq. Mesenchymal clones of mPDAC 021 and mPDAC 901 show an upregulation of genes involved in “MAPK signaling pathway” and “EMT” as compared to the corresponding epithelial clones, in line with increased KrasG12D gene dosage (a full list of enriched gene sets is provided for comparisons in Supplementary Table 15). FDR-adjusted P-values are shown on y axis. Representative data from one experiment are shown. e-g, induction of EMT-like transcriptional programs by KRASG12D overexpression in human PDAC cell lines. e, Graphic of experimental workflow. Two human PDAC cell lines (HUPT3 and PANC0327) with homozygous CKDN2A loss (CDKN2AΔHOM) and heterozygous KRASMUT (KRASMUT-HET) status were transduced with lentivirus carrying doxycycline-inducible KRASG12D or GFP-control expression constructs. KRASG12D or GFP expression was induced by adding doxycycline for 1, 3 and 5 days. f, Gene set enrichment analysis using “Molecular Signatures Database” (MSigDB) of differentially regulated genes in KRASG12D- versus GFP-induced hPDAC cell lines HUPT3 and PANC0327 based on RNA-Seq. Upon doxycycline treatment, both hPDAC cell lines showed a consistent upregulation of genes involved in “KRAS signaling up” and “EMT” (a full list of enriched gene sets is provided for both cell lines in Supplementary Table 16). FDR-adjusted P-values are shown on y axis. g, Expression of marker genes for epithelial (CDH1) or mesenchymal (VIM) cell differentiation and invasion/matrix disassembly (MMP1) was validated by qPCR (normalized to GAPDH and PPIA). In line with RNA-Seq data KRASG12D-induced cells show an increased expression of the mesenchymal marker gene VIM, increased expression of MMP1 and reduced levels of epithelial marker gene CDH1. *P≤0.05, **P≤0.005, ns=not significant, two-tailed t-test; bars=mean; error bars=SEM.