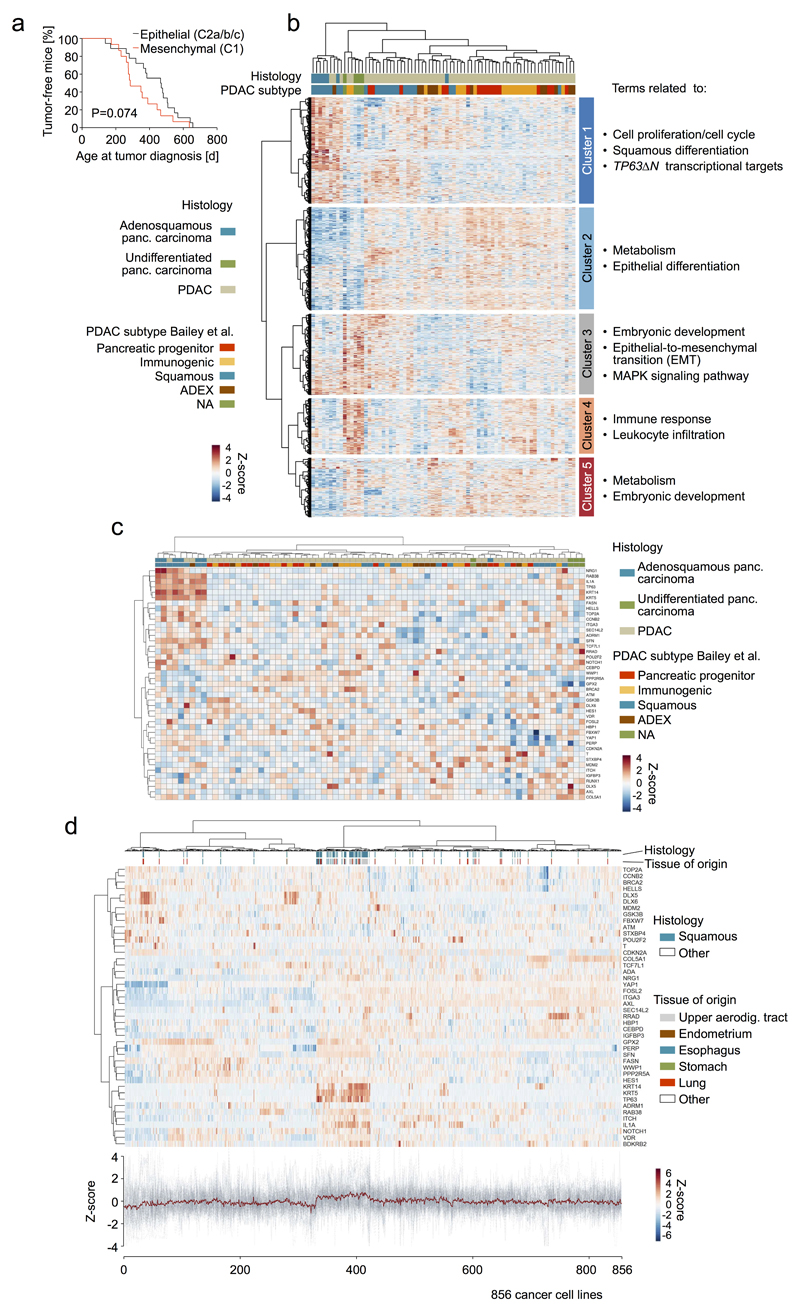
Extended Data Figure 9. Transcriptional profiles of human undifferentiated pancreatic carcinomas are enriched for signatures of oncogenic signaling intensification and EMT but not for activation of TP63ΔN transcriptional network.
a, Primary pancreatic tumors from PK mice with a mesenchymal phenotype (C1 cluster, n=15) are almost exclusively classified as undifferentiated/sarcomatoid by histopathological evaluation and tend to have a reduced age at diagnosis when compared to epithelial (C2a/b/c cluster, n=18) tumors (histopathological grade 1 to 3 [G1-G3]). This aggressive behavior of undifferentiated pancreatic carcinoma is also observed in human patients and is associated with worse clinical outcome33. P-value calculated by two-sided log-rank test. b, Comparison of publically available expression profiles of human undifferentiated pancreatic carcinoma (n=4), PDAC (WHO grade 1 to 3 [G1-G3], n=64) and adenosquamous pancreatic carcinoma (n=7). Human samples with the above histopathological characteristics for which expression-based subtype information from Bailey et al.7 was available were used and complemented with available undifferentiated pancreatic carcinomas from the ICGC PACA-AU cohort (Supplementary Table 18). Other histological subentities of pancreatic cancer were excluded (e.g. IPMN, MCN, acinar cell carcinoma). ANOVA was performed to select genes which are differentially expressed in at least one of the six defined subgroups of pancreatic cancer: (i) undifferentiated, (ii) adenosquamous pancreatic carcinoma and (iii-vi) PDAC (G1-G3) sub-stratified in pancreatic progenitor, immunogenic, squamous and aberrantly differentiated endocrine exocrine (ADEX) Bailey subtypes. Differentially regulated genes were used for unbiased hierarchical clustering of these pancreatic cancer transcriptional profiles. Five sub-clusters of co-regulated gene expression could be identified according to the cluster tree on the y-axis (separated by white horizontal bars in the heatmap). Gene set enrichment analysis using “Molecular Signatures Database” (MSigDB) was performed for individual sub-clusters and terms related to predominating gene sets/pathways are annotated for each cluster on the right (full list provided in Supplementary Table 17). Undifferentiated pancreatic carcinomas cluster together and are associated with (i) upregulation of genes in cluster 3 (containing MAPK signaling pathway and gene sets relevant during embryonic development or EMT) and (ii) downregulation of genes in clusters 2 and 5, which contain gene sets related to epithelial cell differentiation, embryonic development or metabolic signatures. This reflects the pathway enrichment signature in the equivalent undifferentiated (mesenchymal) mouse PDACs (cluster C1/"M" in PK mice; see Extended Data Figure 7g) and provides further support for the link between KRAS signaling intensification, EMT and the undifferentiated tumor phenotype. The immunogenic PDAC subtype showed high expression of cluster 4 genes, which was also strong (even elevated) in undifferentiated pancreatic carcinomas, suggesting an increased immune cell infiltration in undifferentiated carcinomas. Cluster 1 contained gene sets related to cell proliferation/cell cycle, squamous differentiation and TP63ΔN transcriptional targets, which were most highly overexpressed in pancreatic carcinomas with adenosquamous histology. Undifferentiated pancreatic carcinomas did not show activation of the TP63ΔN transcriptional targets. This suggests that activation of TP63ΔN transcriptional targets is not causally linked to KRAS signaling intensification and EMT (see also Extended Data Figure 9c-d, showing a lack of association of undifferentiated carcinomas withTP63ΔN transcriptional network activation). c, Unbiased hierarchical clustering of human pancreatic carcinomas with adenosquamous histology (n=7) as well as PDACs (WHO grade 1 to 3 [G1-G3], n=64) and undifferentiated pancreatic carcinomas (n=4) (sample set as in Extended Data Figure 9b) using a list of validated TP63ΔN transcriptional targets53. Pancreatic cancers with adenosquamous differentiation were significantly enriched in a cluster showing increased TP63ΔN transcriptional network activity (P≤0.001, two-sided Fisher’s exact test, OR 130, 95% CI 11.6-1452). Undifferentiated pancreatic carcinomas did not contribute to this cluster. In line, pancreatic cancers from PK mice did not show differential regulation of the TP63ΔN network, reflecting the lack of adenosquamous tumors in this cohort (not shown). d, Unbiased hierarchical clustering across solid cancers (Cancer Cell Line Encyclopedia, n=856) using the same gene list showed a strong enrichment of tumors with squamous differentiation in the sub-cluster with highest TP63ΔN transcriptional network expression (P≤0.001, two-sided Fisher’s exact test, OR 28.1, 95% CI 16.4-48.1), in line with the observation of Hoadley et al.63 that TP63ΔN is a signature for squamous differentiation across cancers.