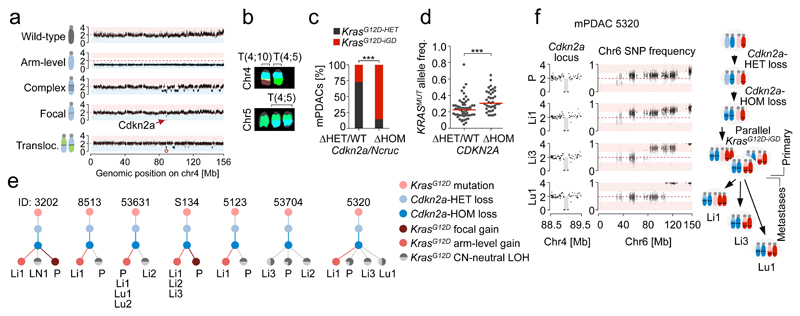
Figure 3. Cdkn2a alteration “states” dictate distinct evolutionary PDAC trajectories.
a, Chr4 alteration types involving Cdkn2a by aCGH/M-FISH (n=38 PK mice). Complex rearrangements, n≥10 CNAs/chromosome. Examplary CNV plots on the right; y-axis, copy number. b, Translocations affecting chr4/Cdkn2a in mPDAC-R1035 by M-FISH (10/10 karyotypes). c, Prevalence of KrasG12D-iGD in mPDAC with homozygously (ΔHOM, n=27) vs. wild-type/heterozygously (ΔHET/WT, n=11) deleted Cdkn2a/Ncruc. ***P=0.001, two-sided Fisher’s exact test, OR 15.3, 95% CI 2.8-83.9. d, KRAS variant allele frequencies in human PDAC with wild-type/heterozygously (n=56) vs. homozygously deleted (n=38) CDKN2A. Data from6. ***P≤0.001, two-sided Mann-Whitney test; bars, median. e, Sequential order of Cdkn2a and KrasG12D alterations. Chr4 and chr6 CNA/LOH patterns (based on aCGH,WES) of primary mPDACs (n=13 PK mice) and associated metastases (n=25). For seven mPDACs and 16 associated metastases the order of genetic events (dots) could be reconstructed. Bifurcations, divergent evolution of clones; lines, lengths do not represent evolutionary distances; P, primary tumor; Li/Lu/LN, liver/lung/lymph node metastasis. f, Detailed chr4/chr6 CNV/LOH profiles for mPDAC5320 primary/metastases. Cdkn2a deletions are identical in all lesions (y-axis, copy number). SNP frequency analysis by WES shows distinct chr6 SNP patterns in metastases and a composite picture in the primary, showing convergent evolution of different KrasG12D-iGD-gains upon Cdkn2aΔHOM. Scheme, combined interpretation of WES/aCGH data.