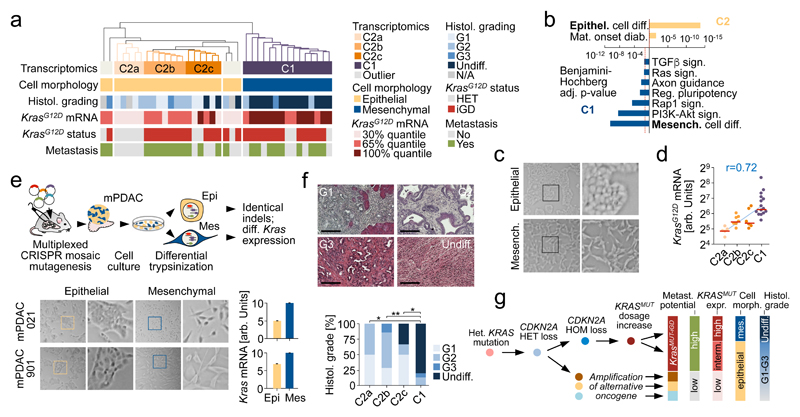
Figure 5. Integrative analyses of PDAC genomics, transcriptomics, cellular phenotypes and histopathologies link molecular, morphologic and clinical disease characteristics.
a, Unbiased hierarchical clustering of primary mPDAC culture transcriptomes (PK mice). Cell morphology, histopathological grading, KrasG12D mRNA expression, genetic KrasG12D status and presence/absence of metastasis integrated below. b, Selected gene sets from gene-set enrichment analysis of clusters C2 vs. C1. (full list in Supplementary Table 13,14). c, mPDAC cultures with mesenchymal/epithelial morphology from clusters C1/C2, respectively. 100x magnification; squares, zoom-in area. d, KrasG12D-allele-specific mRNA levels in mPDAC transcriptional clusters, combined amplicon-based RNA-Seq and qRT-PCR (C2a/b/c/C1, n=5/7/6/15 mice). P=1.9*10-6, two-sided Pearson correlation; bars, median. e, CRISPR/Cas9-mediated multiplexed somatic inactivation of PDAC-relevant tumor suppressors by electroporation-based transfection to achieve low-frequency mosaicism and clonal tumor outgrowth. Differential trypsinization separates epithelial/mesenchymal cells in mPDACs with mixed morphologies (100x magnification; squares, zoom-in area). CRISPR/Cas9-induced indel signatures are identical in epithelial/mesenchymal pairs (Extended data Fig. 8), indicating common cell of origin. Total Kras mRNA levels in epithelial/mesenchymal pairs (qRT-PCR, normalized to Gapdh, n=2 technical replicates). Bars, mean; error bars, SEM. f, mPDAC histophathological grading in transcriptional clusters (C2a/b/c/C1, n=4/7/6/15, single section per mPDAC). Representative sections (H&E) shown. *Benjamini-Hochberg-adj. P≤0.05, **P=0.005; two-sided Fisher’s exact test; scale bars, 150µm. g, Simplified model of PDAC evolution reconciling molecular, morphologic and clinical disease characteristics. KRASG12D-iGD gain or alternative oncogenic amplifications (Myc/Yap1/Nfkb2) are critical for early disease progression. Different oncogenic gains and dosages evolve along distinct evolutionary routes, licensed by defined allelic states (heterozygous/homozygous) and/or combinations of hallmark tumor-suppressor alterations. For simplicity, only the prototype tumor suppressor gene CDKN2A is shown. Not visualized: TP53ΔHOM loss, also promoting KRASMUT-iGD, or TGFBR2ΔHET/HOM inactivation, supporting evolution through CDKN2AHET/KRASMUT-HET trajectories. Depicted alternative trajectories are typical, but not completely exclusive, e.g. MYC or NFKB2 amplifications, which drive KRASMUT-HET cancers, can also cooperate with KRASMUT-iGD. Major aspects of a cancer´s biology/phenotype are linked to differential evolution.