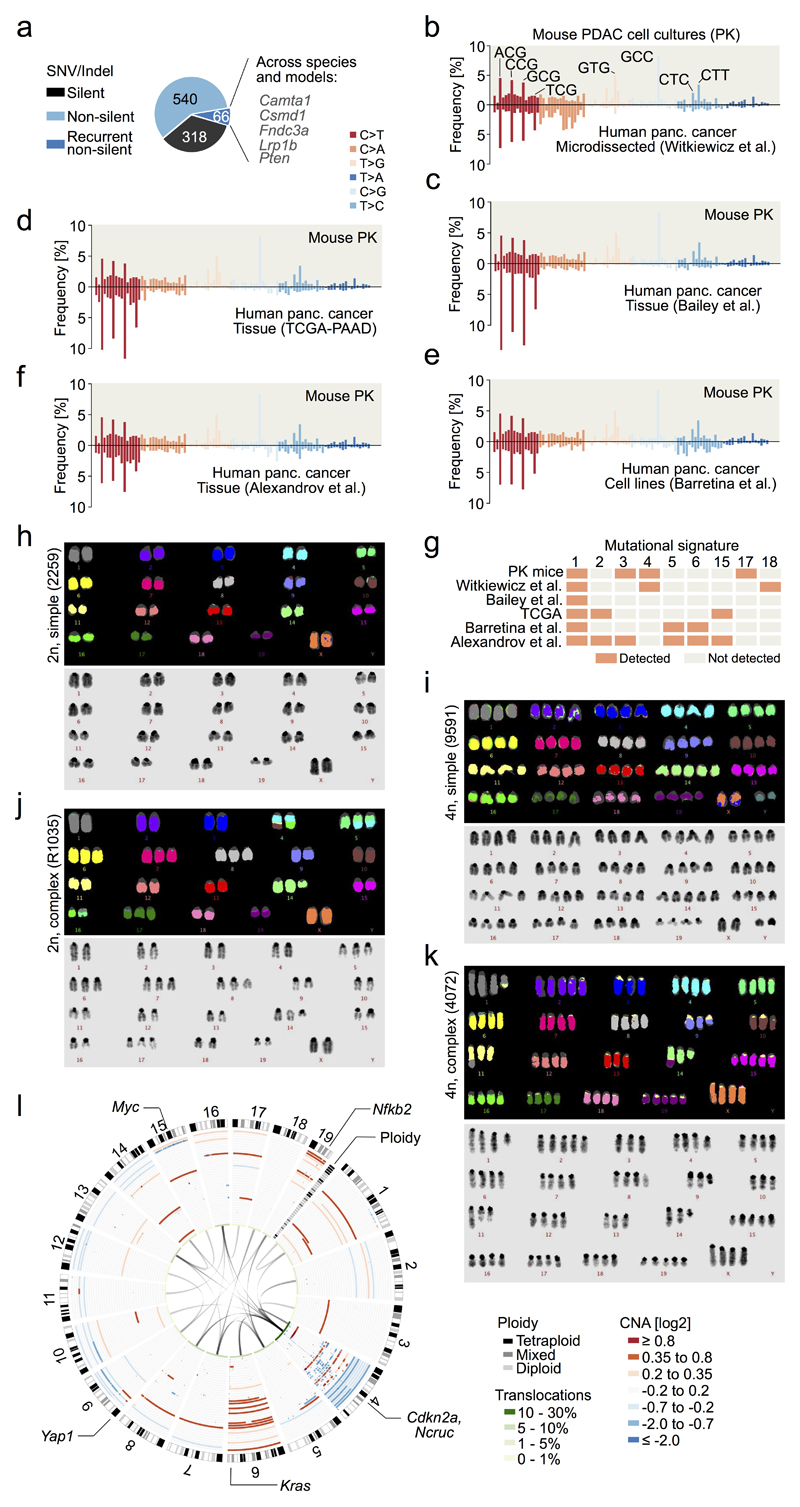
Extended Data Figure 1. Mutational patterns, karyotype complexity and structural alterations in primary PDAC.
a, Single nucleotide variants (SNVs) and indels in primary PDAC cultures derived from 38 KrasG12D (PK) mice, as detected by whole-exome sequencing. Recurrently mutated genes that are frequently altered in human cancers and/or genomewide pancreas-specific transposon screens are indicated. b, Frequency of somatic base substitutions based on trinucleotide context in mouse (n=38 PK mice) and human PDAC (n=51 patients, data used for analysis from 6). b-f, Mutation spectra defined by trinucleotide contexts around base substitutions as detected by whole-exome sequencing show similar patterns in PK mice (n=38) and in relevant human pancreatic cancer cohorts. Base substitutions were extracted from BAM, VCF or MAF files from: b, Witkiewicz et al.6, c, Bailey et al.7, d, TCGA-PAAD, e, Barretina et al.46 and f, Alexandrov et al.47. Additional information regarding the analysis of each cohort is provided in Supplementary Table 2. g, Mutational signatures in mouse and human pancreatic cancer cohorts. Information on mutational signatures was used from Alexandrov et al.47, who identified 21 mutational signatures operative in human cancer. The „deconstructSigs“ tool was used to determine the composition of the given set of 21 mutational signatures in each pancreatic cancer cohort. Extraction of mutational signatures strongly depends on SNV load per tumor. Due to the low mutational burden of mPDACs from PK mice (median of 18 SNVs per tumor as detected by WES), the analyses of mutational signatures could not be performed at the level of individual tumors. We have therefore investigated the contribution of each of the 21 mutational signatures to the SNV spectrum at the cohort-level (see Methods). Signature 1, reflecting age-associated C>T transversions at NCG trinucleotides, was the only signature consistently identifiable in all cohorts of human and mouse pancreatic cancer. In comparison to human cohorts, PK mice show C>G substitutions at GCC trinucleotides that cannot be attributed to one of 21 mutational signatures. Note that mutations at the GCC motif are not a general phenomenon of PDAC from PK mice, since only 4 samples are predominantly contributing to this peak. h-i, Representative M-FISH karyotypes with no or few karyotypic changes are shown for a diploid (40 chromosomes) and tetraploid mouse PDAC (81 chromosomes). Tumor 9591 shows gain of chr14. j, Representative karyotype of a complex diploid mPDAC genome with aneuploidy and translocations (46 chromosomes). Both copies of chr4 are involved in translocations: der(4)t(4;10) and der(4)t(4;16); likely affecting Cdkn2a. Further structural alterations and copy number changes are: +5, der(5)t(4;5)*2, +6, +7, +8, del(9), +14, del(14), der(16)t(5;16), +17. k, Representative example of a complex tetraploid mPDAC karyotype (77 chromosomes). Structural alterations are: der(1)t(1;11), dic(9;9), der(11)t(1;11), and der(14)t(14;19). Single chromosomal copy number changes are: +2, -3, -9, -10, -11, -13, -14, +15 and +19. Del, deletion; der, derivative chromosome; dic, dicentric chromosome; t, translocation; „-“, chromosome loss; „+“, chromosome gain. l, (Extension to Fig. 1c.) Circos plot shows CNAs assessed by aCGH as well as translocations and ploidy states detected by M-FISH in 38 primary PDACs derived from PK mice (n=38). CNAs for each mPDAC are displayed as log2 difference from tail control. Frequencies of translocations per chromosome are indicated in green in the inner circle of the graph. Connecting lines indicate individual translocations and involved chromosomes. On chr4, genomic alterations frequently involve Cdkn2a or Ncruc, a Non-coding regulatory region upstream of Cdkn2a (27/38 cancers with homozygous and 10/38 with heterozygous inactivation of Cdkn2a and/or Ncruc). Only one cancer remained Cdkn2aWT. The target of copy number changes on chr6 is KrasG12D, either through arm level gain or focal amplification. In addition, primary mPDAC of PK mice exhibited recurrent genetic amplifications affecting other known oncogenes, such as Myc or Yap1, or Nfkb2, a novel oncogenic PDAC driver identified in this study (see also Fig. 2e,f and Extended Data Figure 4).