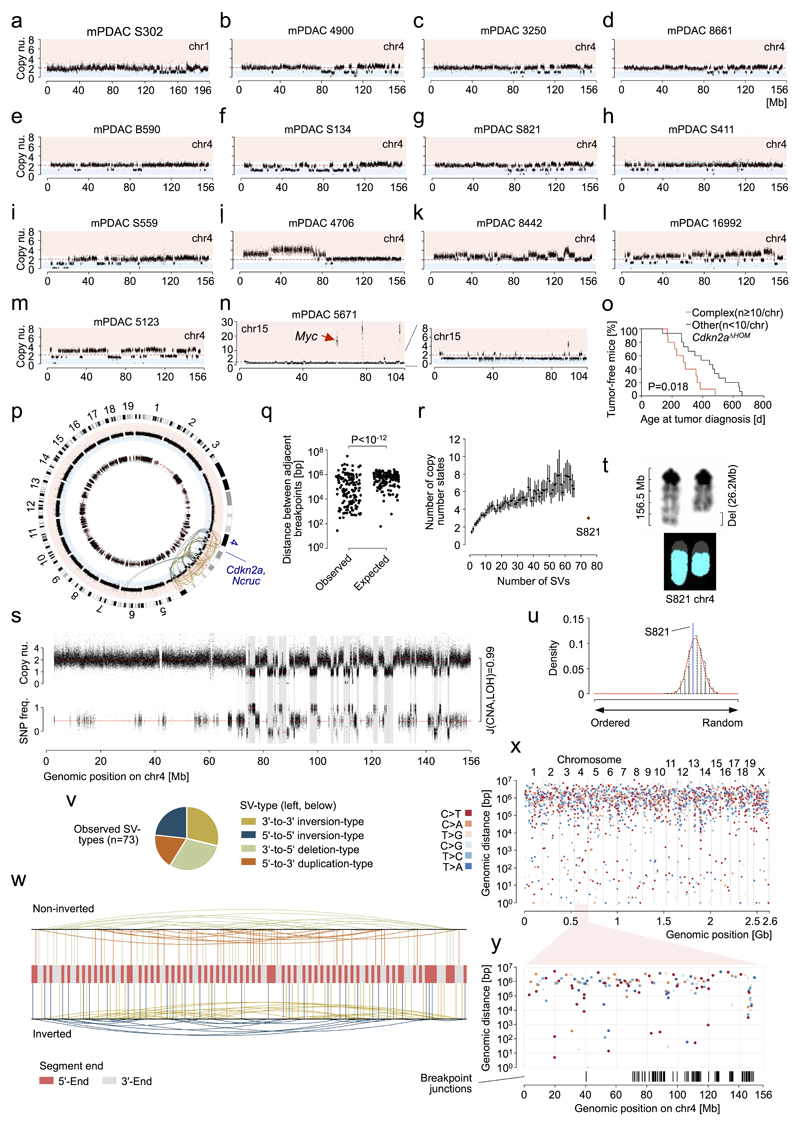
Extended Data Figure 2. Characterization of complex rearrangements in PDAC from PK mice and statistical inference of chromothripsis based on whole genome sequencing (WGS).
a-n, Copy number profiles of chromosomes with complex rearrangements (defined as n≥10 CNAs per chromosome) from primary mPDAC cell cultures as detected by aCGH. A total of 14 mPDACs had chromosomes with complex rearrangements. a-i, Nine primary mPDACs show copy number patterns characterized by heterozygous deletions and oscillation of copy number around few states, indicating chromothripsis as the underlying mechanism. g, mPDAC-S821 was subjected to whole genome sequencing for the inference of chromothripsis using previously established criteria14 (see Fig. 1d and Extended Data Figure 2p-w). j-m, Four primary mPDACs showed complex rearrangements with multiple copy number states on chr4, likely acquired through progressive/sequential rearrangement cycles. n, Cancer 5671 carries a complex rearrangement on chr15 characterized by oscillating copy number states and 3 prominent focal amplifications, of which one contained the Myc oncogene. Myc amplification is most likely the result of double minute chromosome formation during chromothriptic rearrangement of chr15. o, Comparison of age at tumor diagnosis in Cdkn2aΔHOM-deleted cancers with (n=10) or without (n=15) complex clustered chromosomal rearrangements (n≥10 CNAs/chromosome). Complex clustered rearrangements are associated with significantly shortened time to tumor diagnosis, indicating accelerated tumor evolution through genetic crisis. Two-sided log-rank test. p, Criteria proposed by Korbel et al.14 were tested for the inference of chromothripsis. Circos plot displays SNP ratio (inner circle, red dashed line indicating heterozygosity), CNV (outer circle, blue area indicating deletion, red amplification) and structural variations (SVs, colors as in v) as detected by WGS. Chr4 shows a complex deletion pattern and massive rearrangements associated with loss of one copy of Cdkn2a. The second copy of Cdkn2a is focally deleted. In addition, a balanced translocation of a ~200Kb segment from trisomic chr6 to chr4 and a far smaller segment of chr4 into chr6 was detected. The Kras locus is not directly affected by this inter-chromosomal translocation. LOH, CNAs and rearrangements are not detected on other chromosomes. q, In a chromothriptic model, DNA breakpoints tend to cluster on a chromosome. Testing against an exponential distribution (parameter λ derived from mean of observed distance between adjacent breakpoints), revealed significantly shorter distances than expected in a progressive model (n=146 breakpoints). P<10-12; χ_-goodness-of-fit test. r, In a progressive model of acquisition of massive rearrangements, copy number states tend to be more complex than in the chromothriptic. Monte Carlo simulations were used to generate a progressive evolution model with sequential accumulation of observed rearrangements (n=100 simulations per number of SVs). mPDAC S821 showed fewer copy number states on chr4 than expected in the progressive model. Mean is indicated as a black point and lines represent the 95% CI. s, Chromothriptic tumors typically feature interspersed loss and retention of heterozygosity. Accordingly, there was a high overlap between deleted regions and LOH segments on chr4 (Jaccard index (J) = 0.99). t, In a chromothriptic model, DNA shattering typically occurs on a single haplotype. M-FISH showed that significant loss of chromosomal content occurred on only one copy of chr4. u, To show random chromothriptic DNA shattering and re-joining, observed segments (n=73) were re-ordered by running Monte Carlo simulations (n=103) that generate a background probability distribution. S821 segment order lies within the chromothriptic null model. Two-sided P=0.78. v, All 4 SV-types are uniformly distributed in a chromothriptic tumor model. P=0.43; χ_-goodness-of-fit test. w, In a chromothriptic model, paired end connection types (as given by the SV-type) induce an alternating sequence of DNA segment ends when ordered according to the genomic position on the original chromosome. Tendency towards this alternating 3’-to-5’ pattern of rearranged DNA segment ends (n=146) was tested by using right-sided Wald-Wolfowitz runs test. P<10-12. x, Mutation clusters in relation to breakpoint junctions involved in chromothripsis are shown as rainfall plot for primary PDAC from PK mouse S821. Each dot represents a single somatic nucleotide variation (SNV) and is ordered on the x-axis according to its position in the mouse genome. The distance of each SNV to the previous SNV in the genome is shown on the y-axis. The coloring of individual SNV dots indicates the type of nucleotide substitution. y, Chr4 “zoom-in” from (x). Breakpoint junctions are shown according to their genomic position on chr4. No mutation clusters - neither in absence nor in combination with breakpoint junctions - were detected, consistent with chromothripsis involving end joining DNA repair mechanisms. This is in contrast to other complex rearrangement types, such as chromoanasynthesis, which arise through replication-based mechanisms with breakpoint-associated high mutation rates (e.g. kataegis).