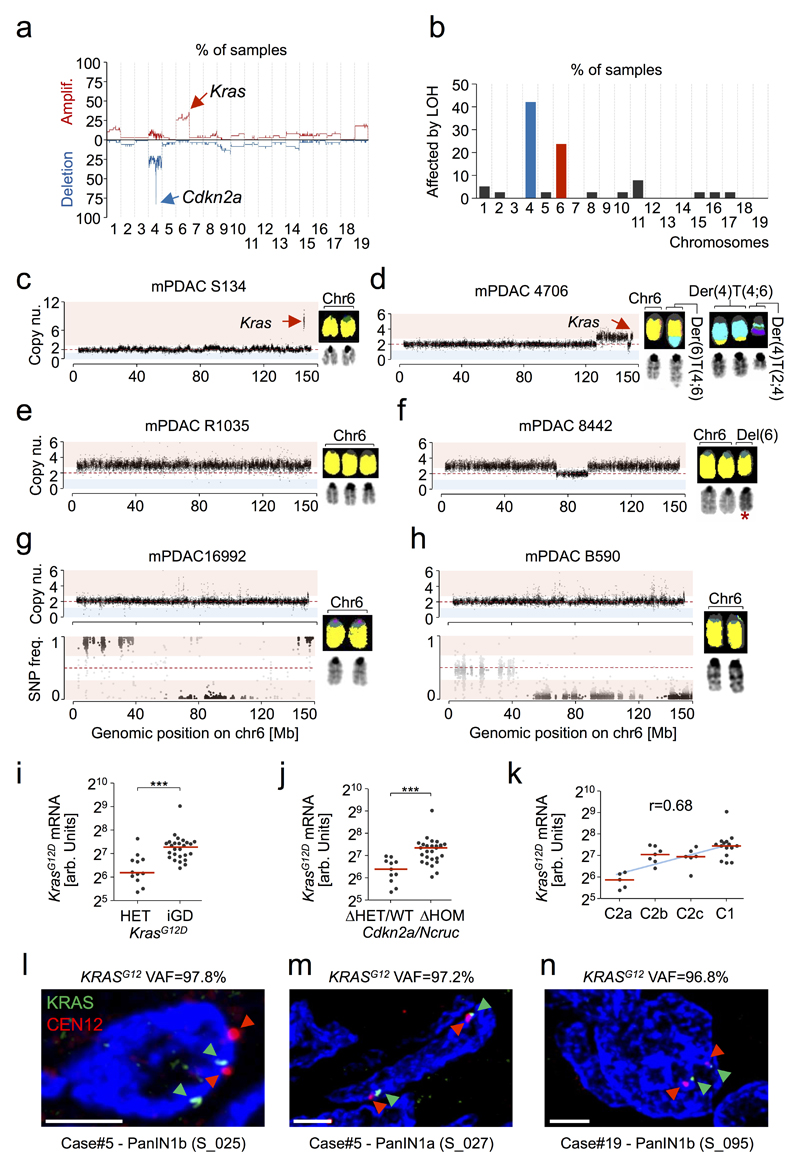
Extended Data Figure 3. Specificity, timing, mechanisms and impact of KrasG12D gene dosage alterations on gene expression in pancreatic tumorigenesis.
a, Overlay of copy number profiles of primary mPDAC cell cultures from PK mice (n=38) as determined by aCGH. Y-axis shows frequency of a genomic region to be amplified (up) or deleted (down) in the cohort, with Cdkn2a and Kras loci being most frequently affected by CNAs. b, Prevalence of LOH in primary mPDAC cell cultures from PK mice (n=38) based on whole exome sequencing (WES) data. A chromosome was considered to be affected by LOH if the SNP frequency was shifted to ≤0.1 or ≥0.9 in a segment with a size ≥200kb. LOH on chr4 is frequently the consequence of heterozygous deletions involving the Cdkn2a locus. By contrast, LOH on chr6 is predominantly copy number neutral and linked to increase in KrasG12D gene dosage. Chr4 (home of Cdkn2a) and chr6 (home of Kras) show markedly increased rates of LOH as compared to all other chromosomes reflecting their functional importance during tumorigenesis. c-h, Genetic mechanisms of KrasG12D gene dosage alterations as identified by aCGH, M-FISH and whole exome sequencing (WES) in pancreatic cancers from PK mice. The observed types of increased KrasG12D gene dosage acquisition were: (i) focal gain (affecting ≤50% of the chromosome length), arising either through replication-based mechanisms (2 cases, one with high-level KrasG12D amplification [shown in c] and one with low level amplification) or translocation and subsequent amplification of the translocated chromosome (one case [shown in d]), (ii) arm-level gain (affecting ≥50% of the chromosome length) arising through mitotic errors (7 cases of whole-chromosome gain [example shown in e], occasionally [2 cases] with concomitant intra-chromosomal deletions or translocations not affecting Kras [example shown in f]) and (iii) copy-number neutral LOH (CN-LOH, KrasG12D homozygosity, acquired uniparental disomy), arising either through mitotic recombination (affecting parts of chr6 [shown in h]) or chromosomal missegregation (duplication of KrasG12D-mutant chr6 and loss of wild-type chr6 [shown in g]). c, mPDAC S134 shows a high-order focal amplification of KrasG12D. Sharp borders, small size of the amplification (600kb) and strong increase in copy number (4x) indicate that KrasG12D was amplified through multiple cycles of repeated template-switching by a replication-based DNA repair mechanism. KrasG12D mutant allele frequency is 89.1%. d, Tumor 4706 carries a focal amplification of KrasG12D. M-FISH analysis revealed that the mutant KrasG12D allele (chr6) was likely first affected by a reciprocal translocation of chr4 and chr6, resulting in two rearranged chromosomes: Der(4)T(4;6) and Der(6)T(4;6). Subsequently, Der(4)T(4;6) was missegregated through mitotic error resulting in focal gain of the KrasG12D locus. KrasG12D mutant allele frequency is 72.2%. e, mPDAC R1035 shows ‘classical’ whole chromosome gain (trisomy) of chr6, which was likely generated through mitotic error/missegregation. The KrasG12D mutant allele frequency is 69.8%. f, In tumor 8442 arm-level gain of KrasG12D was likely generated through mitotic missegregation of chr6. Intra-chromosomal deletion on one of three chromosomes (19.6Mb) does not affect Kras. KrasG12D mutant allele frequency is 66.4%. Asterisk, chr6 with reduced length resulting from intra-chromosomal deletion. g-h, mPDAC 16992 and B590 display copy-number neutral LOH (CN-LOH) leading to increased KrasG12D gene dosage. KrasG12D mutant allele frequency is 99.2% and 96.3%, respectively. The SNP pattern of chr6 in mPDAC 16992 reveals that the whole chromosome is affected by CN-LOH indicating chromosome missegregation (duplication of the KrasG12D-mutant chr6 and loss of wild-type chr6) as the underlying mechanism. By contrast, in mPDAC B590 only a partial region of chr6 is affected by CN-LOH, therefore probably resulting from mitotic recombination. i, Allele-specific KrasG12D mRNA expression in KrasG12D-HET (n=12) vs. KrasG12D-iGD (n=26) primary PDAC cell cultures from PK mice as detected by combined analysis of amplicon-based RNA-Seq (proportion of mutant/wild-type Kras mRNA) and 3’-prime pA RNA-Seq (amount of total Kras mRNA, but not the proportion of mutant/wild-type Kras mRNA due to sequencing of 3’-prime transcript ends; see Methods section). This figure is related to Fig. 2b. ***P≤0.001, two-tailed Mann-Whitney test; bars, median. j, Mutant KrasG12D mRNA levels in Cdkn2a/NcrucΔHET/WT (n=11) vs. Ckdn2a/NcrucΔHOM (n=27) primary PDAC cell cultures from PK mice as detected by combined amplicon-based RNA-Seq and 3’-prime pA RNA-Seq. This figure is related to Extended Data Figure 5f. ***P≤0.001, two-tailed Mann-Whitney test; bars, median. k, Mutant KrasG12D mRNA levels in transcriptional clusters of mPDAC from PK mice (C2a/b/c/C1, n=5/7/6/15) as detected by combined amplicon-based RNA-Seq and 3’-prime pA RNA-Seq. This figure is related to Fig. 5d. P=1.6*10-5, two-sided Pearson correlation; bars, median. l-n, Interphase fluorescence in situ hybridization (FISH) for the analysis of copy-number and ploidy states at the KRAS locus on chr12 in human pancreatic intra-epithelial neoplasia (PanIN) with KRASG12 variant allele frequencies (VAFs) of ~100%. KRASG12 VAFs are indicated above each FISH profile as detected by amplicon-based deep sequencing. A VAF of ~100% can be caused either by loss of the wild-type KRAS-locus (hemizygosity of KRASG12-MUT: one KRASG12-MUT allele per cell) or by CN-LOH (acquired uniparental disomy; homozygosity of KRASG12-MUT: two KRASG12-MUT alleles per cell). All samples show a diploid genome as suggested by CEN12 (two red signals per nucleus). Neither loss of one KRAS allele nor monosomy of chr12 was observed providing evidence for CN-LOH and increased KRASG12-MUT gene dosage in hPanIN. Scale bars, 2.5µm; CEN12, centromere probe chr12.