Figure 5. TIN2 Blocks the Fbx4-TRF1 Interaction and Inhibits the Ubiquitination of TRF1 Mediated by SCFFbx4.
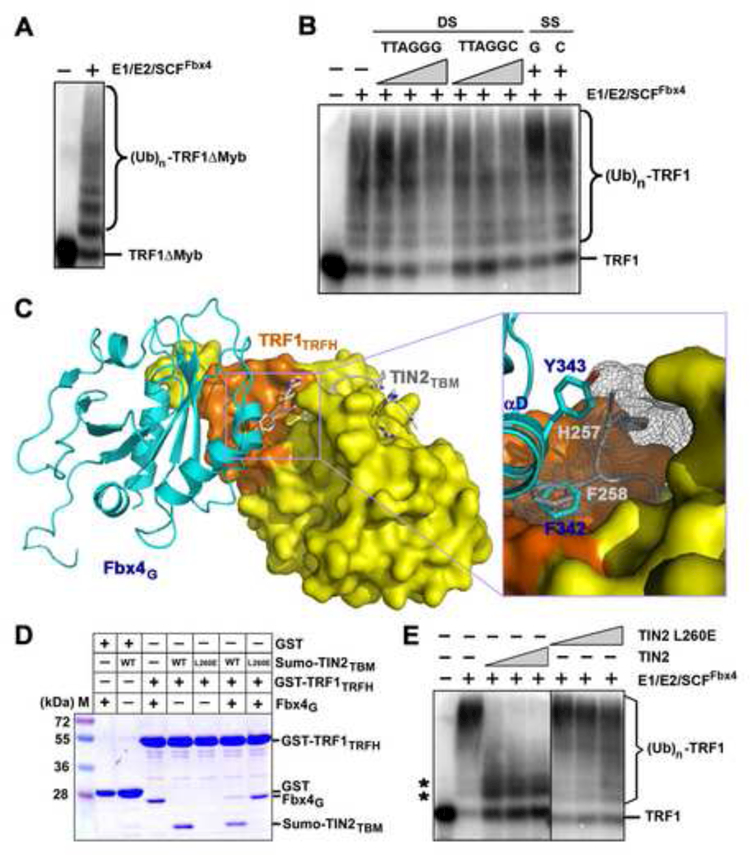
(A) TRF1 deletion mutant lacking the DNA-binding Myb domain, TRF1ΔMyb, can be efficiently ubiquitinated by SCFFbx4.
(B) Addition of either wt or mutant telomere DNAs has no effect on TRF1 ubiquitination. In vitro ubiquitination reactions containing 33P-labeled TRF1 were carried out in the absence (−) or presence of double-stranded (DS; 1, 12 or 36 μM TTAGGG or TTAGGC) repeat DNA or single-stranded [SS; 36 μM TTAGGG (G) or AATCCC (C)] repeat DNA.
(C) Superposition of the Fbx4G-TRF1TRFH structure with the crystal structure of the TRF1TRFH-TIN2TBM complex. Fbx4G (cyan) and TIN2TBM (gray) share a common hydrophobic interacting surface on TRF1TRFH (orange). An enlarged view of the collision between Fbx4G αD helix and the TIN2TBM peptide is highlighted in the box.
(D) GST pull-down competition assay of the Fbx4G-TRF1TRFH interaction in the presence of wt or the L260E mutant TIN2TBM peptides. For clarity, Sumo-fused TIN2TBM peptides were used in this assay. Sumo itself does not bind to TRF1TRFH (data not shown).
(E) TIN2 inhibits Fbx4 mediated ubiquitination of TRF1. TRF1 ubiquitination assays were performed as in (A), but increasing amount of TIN2 (3, 9, 27, 81 μM) or mutant TIN2L260E (3, 9, 27, 81 μM) was added in the reactions. Asterisks indicate the non-specific ubiquitination products of TRF1 due to the presence of Cul1/Rbx1.
