FIGURE 1.
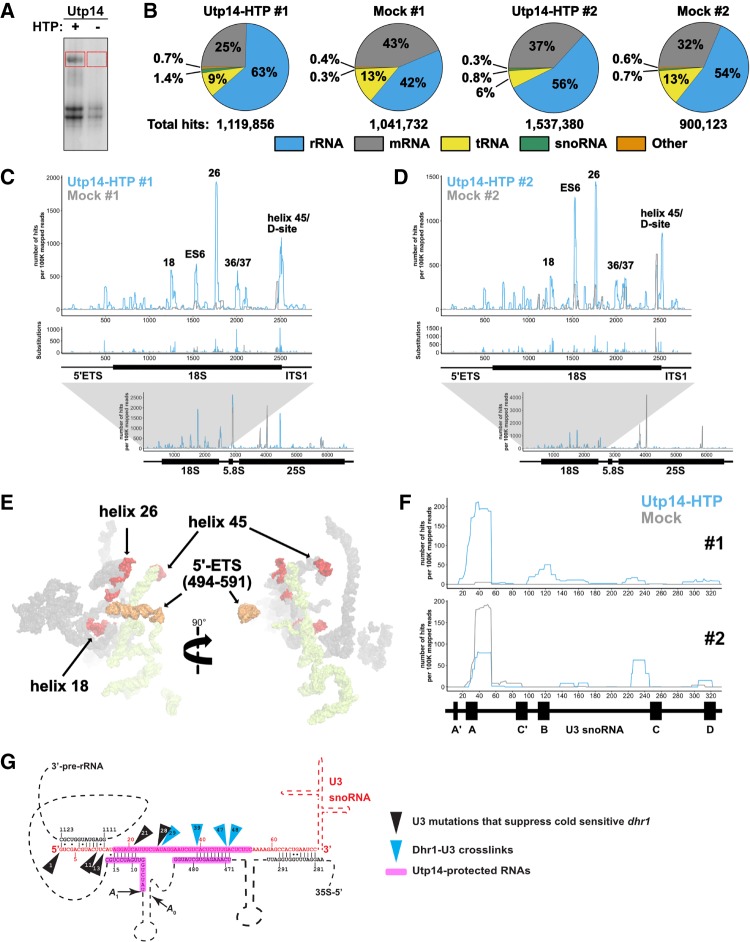
Utp14 crosslinks to multiple regions within the pre-18S rRNA. (A) A representative autoradiograph of 32P-labeled RNAs crosslinked to Utp14-HTP (+, AJY4051) and mock (−, BY4741). Red boxes indicate the regions of the membrane that were excised and used in library preparation. (B) Percentages of RNA composition grouped by class are shown for both CRAC replicates. Total aligned reads corresponding to each sample are shown below. (C,D) The number of hits per 100K mapped reads (top) and substitutions (middle) are shown against nucleotide position within the pre-18S rRNA (RDN18-1). The number of reads against nucleotide position within the 35S pre-rRNA (RDN37-1) are shown below. Utp14-HTP is shown in light blue, and the mock is shown as gray. Two independent biological replicates are shown. (E) Utp14 crosslinks within 18S rRNA (red) and 5′-ETS (orange) mapped to a recent structure of the SSU processome. RNAs that were not crosslinked with Utp14 are shown in surface representation for18S rRNA (gray) and 5′-ETS (yellow) (PDB: 5WYJ). (F) The number of hits per 100K mapped reads against nucleotide position within U3 (snR17A) for data sets #1 (top) and #2 (bottom). A diagram of U3 is shown below the plots. (G) A cartoon of U3 hybridization to the rRNA within the SSU processome displaying Dhr1 crosslinks (blue triangles) and mutations of U3 that suppress a cold-sensitive Dhr1 mutant (black triangles) (Sardana et al. 2015) in relation to Utp14 crosslinks to U3 and the rRNA (magenta highlights). Relevant data processed using pyCRAC are reported in Supplemental File 1.