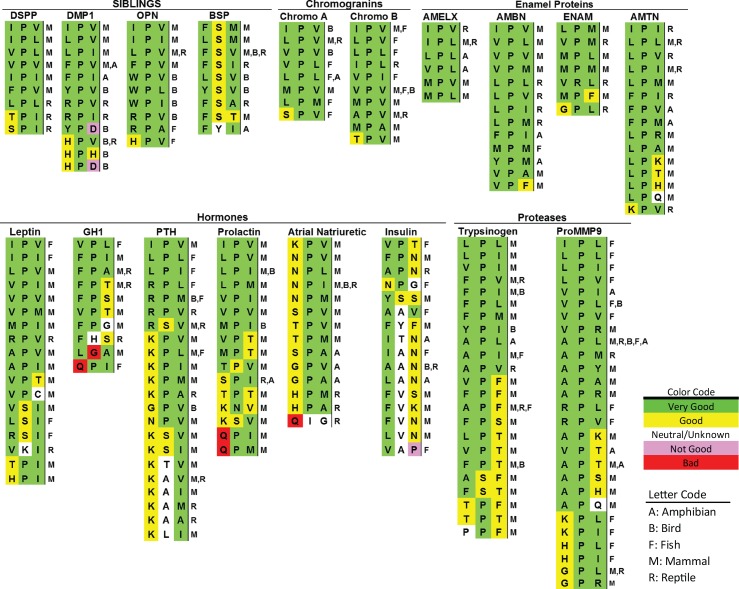
Fig 1. Examples of starting tripeptides for several classes of secreted vertebrate proteins with conserved, ER-ESCAPE motifs.
All tripeptides found in NCBI Protein database searches (by gene name and BLASTP) are listed a single time for each protein, with vertebrate taxon notations in single-letter codes on the right. Signal peptide cleavage sites were predicted by SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/) [6], Phobius (http://phobius.sbc.su.se/) [7], and/or experimental evidence noted on NCBI Proteins database. (See S1 Table for accession number, species name, and brief sequence of representative taxon for each tripeptide.) Color-coding based on relative contribution of each amino acid position to the strength of the ER-ESCAPE motif is as noted in Results and Discussion. AMBN, ameloblastin; AMELX, amelogenin, X-linked; AMTN, amelotin; BSP, bone sialoprotein; DMP1, dentin matrix acidic phosphoprotein 1; DSPP, dentin sialophosphoprotein; ENAM, enamelin; ER-ESCAPE motif, Endoplasmic Reticulum Exit by Soluble Cargo using Amino-terminal Peptide-Encoding motif; GH1, growth hormone 1; NCBI, National Center for Biotechnology Information; OPN, osteopontin; proMMP-9, pro-matrix metalloproteinase-9; PTH, parathyroid hormone.