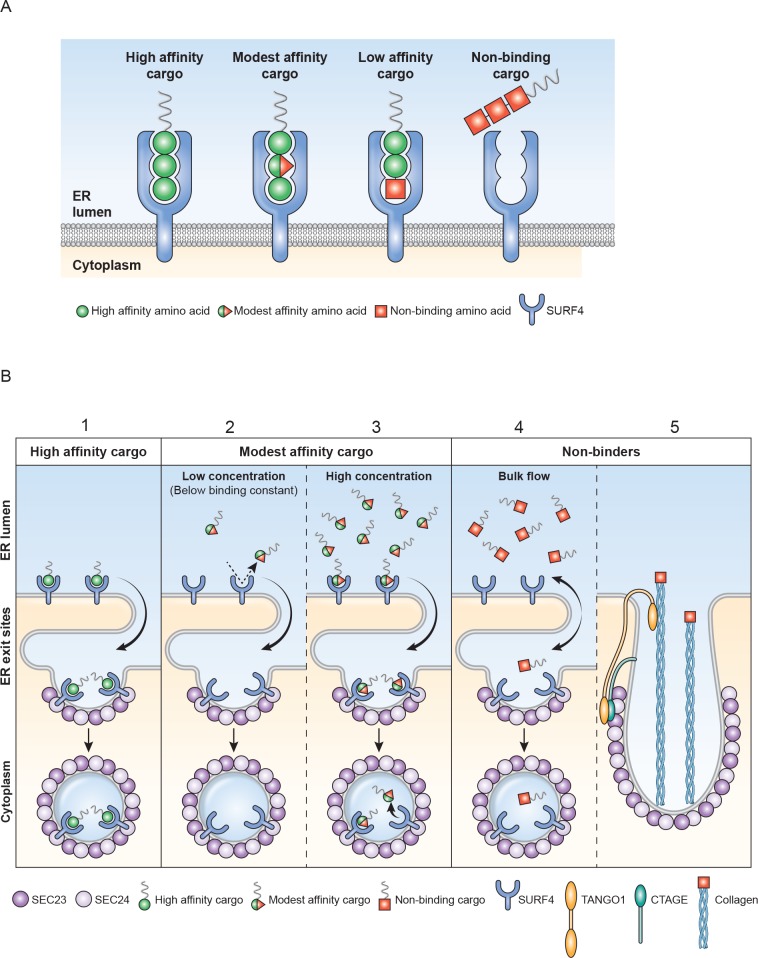
Fig 11. Model illustrating interaction of ER-ESCAPE motifs with high, modest, and no affinity for Surf4/Erv29p.
(A) A green ball amino acid in spherical pocket denotes highest contribution of that residue to binding affinity such as a proline in number 2 position or a hydrophobic residue in position 1 (amino-terminus) or 3. Green half-ball plus red pyramid represents lower affinity interaction for that amino acid (e.g., serine in position 2), while the red cube denotes a negative contribution to binding affinity (e.g., acidic amino acid in position 1). High-affinity cargo present high-affinity contributions in all three positions, while modest- to low-affinity tripeptides have at least one mismatch. Nonbinding proteins such as chaperones or fibrillar collagens have two or three completely mismatching amino acids. (B) (1) High-affinity cargo (e.g., IPV) are bound to Surf4/Erv29p and exit ER before they accumulate to aggregate-forming concentrations. (2) Cargo with more modest ER-ESCAPE motifs (e.g., FSM) do not significantly bind to cargo receptor until (3) they accumulate to levels ≥ their binding constant. Only at that point do they remain bound long enough to remain in COPII vesicle at levels significantly greater than bulk flow. (4) illustrates cargo starting with nonbinding amino-terminal tripeptides (e.g., QEE) cannot exit ER more efficiently than their concentration in ER lumenal fluid in equilibrium with the small amount of exit vesicle fluid (bulk flow). (5) Fibrillar collagens are too large for standard COPII exit vesicles and must use more voluminous TANGO1/cTAGE5-associated exit vesicles. Large fibrillar collagens often start with nonbinding motif (e.g., QEE) to keep them from binding Surf4 and partially entering smaller COPII vesicles. COPII, coat protein complex II; ER, endoplasmic reticulum; ER-ESCAPE motif, ER-Exit by Soluble Cargo using Amino-terminal Peptide-Encoding motif; FSM, phenylalanine-serine-methionine; IPV, isoleucine-proline-valine; QEE, glutamine–glutamic acid–glutamic acid; Surf4, surfeit locus protein 1; TANGO1, transport and Golgi organization 1.