Fig. 5. Covalent bonding of conformational states and higher-order assemblies.
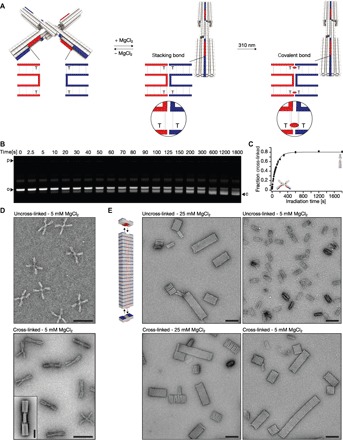
(A) Schematics of the two-state switch that consists of two rigid beams flexibly connected in the middle by an immobile Holliday junction. Cylinders in the models represent double-helical DNA domains, and shape-complementary surface features are highlighted in red and blue. Insets show blow-ups of the blunt-ended interfaces of protruding (red) and recessive (blue) surface features. Thymidines directly located at the blunt-end site can be cross-linked upon UV light irradiation. The resulting CPD bond is indicated as a red ellipsoid. (B) Laser-scanned fluorescent image of 2.0% agarose gel stained with ethidium bromide. Switch samples were irradiated at 310 nm for different periods of time and loaded on the gel. o and c, species of particles populating open and closed state, respectively. (C) Plot of the fraction of cross-linked switch particles as a function of time obtained from the gel in (B). The experiment was performed in triplicate; data points represent the mean, and error bars represent the SD. (D) Exemplary TEM micrographs. Top: Nonirradiated sample with particles populating the open state. Bottom: Irradiated (20 min at 310 nm) sample with particles locked in the closed conformational state. Scale bars, 100 nm. Inset: Average 2D particle micrograph of cross-linked particles. Scale bar, 20 nm. (E) Top left: Model of the multilayer DNA origami brick that polymerizes into linear filaments. Fields of view of TEM micrographs recorded at the indicated conditions. Scale bars, 100 nm.
