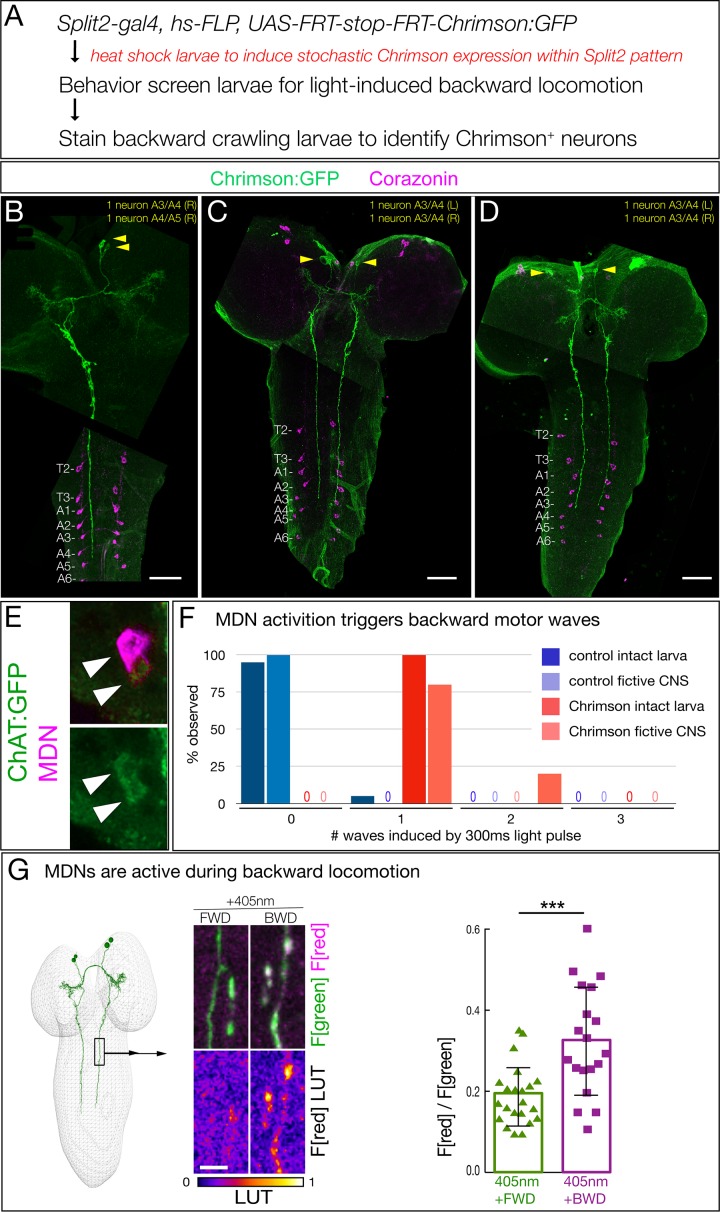
Figure 2. Two brain descending neurons are sufficient to induce backward larval locomotion.
(A) Experimental flow for generating sparse, stochastic patterns of Chrimson in subsets of the Split2 expression pattern. Genotype: hsFlpG5.PEST R49F02-Gal4AD R53F07-Gal4DBD tsh-lexA, lexAop-killer zipper UAS.dsFRT.Chrimson:mVenus. (B–D) The CNS from three larvae that crawled backward in response to Chrimson activation. All show expression in neurons with medial cell bodies, bilateral arbors, and a contralateral descending projection to A3-A5. (B) Note there is a tear in the CNS near segment T1. Chrimson:Venus, green; Corazonin (Crz; segmental marker), magenta. Scale bar, 50 μm. (E) MDNs are cholinergic. MDNs marked with mCherry (magenta) express ChAT:GFP (green). Genotype: R49F02-Gal4AD,UAS-Chrimson:mCherry; R53F07-Gal4DBD, mimic ChAT:GFP. (F) A brief pulse of MDN activity can trigger a backward wave. Intact larvae: individual L3 larvae were subjected to 300 ms of 561 nm light and the number of backward waves was quantified (n = 20). Genotype: ss01613-Gal4 (Split3); UAS-Chrimson::mVenus. Fictive CNS: isolated L3 CNS was subjected to 300 ms 561 nm light and the response of the downstream neuron A18b was monitored for a backward wave of GCaMP6f activity. All MDN activations led to at least one backward wave, but there are also backward waves that occur independently of MDN activation (see Discussion), and these may account for the observed second waves. Genotype: ss01613-Gal4 (Split3)/R94E10-lexA; lexAop-GCaMP6f,UAS-Chrimson:mCherry. (G) MDNs are preferentially active during backward (BWD) not forward (FWD) locomotion in the intact larva. CaMPARI in MDN descending projections within the SEZ of third instar larvae. Top, fluorescence emission following excitation by 488 nm (green) or 561 nm (magenta); bottom, emission from 561 nm imaging alone. Right, quantification of red fluorescence over green fluorescence, mean intensity. Each value represents data from an individual descending projection. See Materials and methods for details. n = 22 for FWD and 19 for BWD. Scale bar, 10 μm. Genotype: R49F02-Gal4AD R53F07-Gal4DBD UAS-CaMPARI.