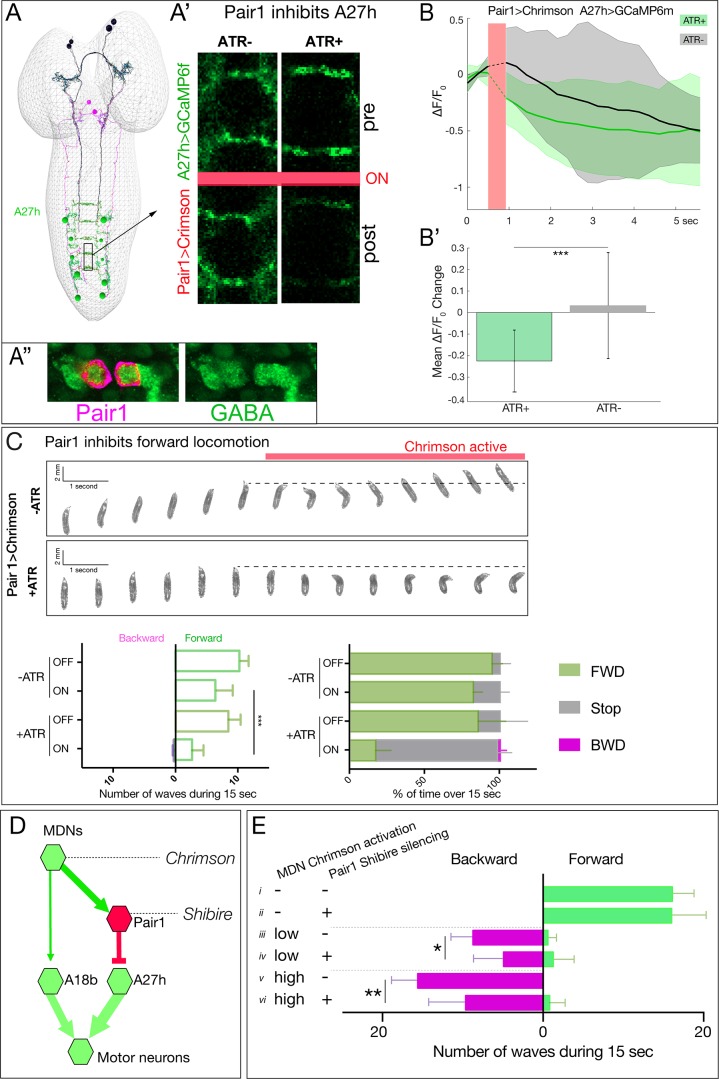
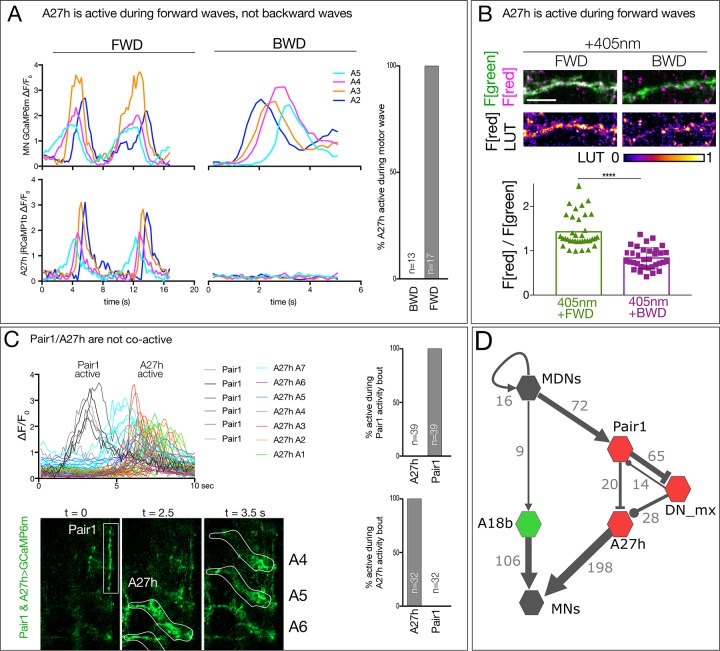
Figure 7. Pair1 inhibits the forward-active A27h premotor neuron, and arrests forward locomotion.
(A,B) Pair1 inhibits A27h. (A) Reconstruction of MDNs (black), Pair1 neurons (magenta) and A27h neurons (green) in the first instar CNS TEM volume. (A’) A27h GCaMP6m fluorescence is reduced following Pair1 Chrimson activation (red bar); two segments shown. (A’’) Pair1 is GABAergic. Pair1 cell body (mCherry; magenta, arrowheads) and GABA (green). Genotype: R75C02-Gal4, UAS-Chrimson:mCherry. (B) A27h GCaMP6m fluorescence is reduced following Pair1 Chrimson activation (red bar). (B’) ΔF/F0 was significantly inhibited in ATR +animals relative to ATR- controls. A total of 26 events from seven animals were averaged for ATR +and 16 events from four animals for ATR- group. See Materials and methods for further details. Genotype: R75C02-lexA/+; lexAop-Chrimson:mCherry/R36 G02-Gal4, UAS-GCaMP6m.. (C) Activation of Pair1 halts FWD locomotion for the duration of neuronal activation. Top, time-lapse images of ±ATR larvae expressing Chrimson in Pair1 neurons before and during light stimulation. Bottom left, backward and forward wave number over 15 s without Chrimson activation (Off) or during Chrimson activation (On) in third instar larvae. n = 12 for all groups. Bottom right, percent time performing forward locomotion (green), backward locomotion (magenta) or not moving (grey) over 15 s without Chrimson activation (Off) or during Chrimson activation (On) in third instar larvae. n = 5 for all groups. Genotype: R75C02-Gal4 UAS-Chrimson:mVenus.. (D) Schematic illustrating the experiment in (E). Arrows, excitatory connections; T-bar, inhibitory connection; line width proportional to synapse number. (E) Pair1 activity is necessary for efficient Chrimson-induced backward locomotion. Chrimson was expressed in MDNs, and shibirets was expressed in Pair1 neurons. Low (0.07 mW/mm2) or high (0.275 mW/mm2) light intensities were used to induce MDN activity; a temperature shift to 32°C was used to inactivate Shibirets and thus silence Pair1 neurons. Silencing of Pair1 alone had no detectable phenotype (i, ii). Silencing Pair1 decreased the efficacy of MDN-induced backward locomotion at low or high light levels (iii-vi). Genotypes: R49F02-Gal4AD R53F07-Gal4DBD UAS-Chrimson:mVenus pBD-lexA lexAop-Shibirets (i, iii and v) and R49F02-Gal4AD R53F07-Gal4DBD UAS-Chrimson:mVenus R75C02-lexA lexAop-Shits1 (ii, iv and vi).