Figure 2.
NUP214-ABL1 and TLX1 Upregulate the JAK-STAT Pathway
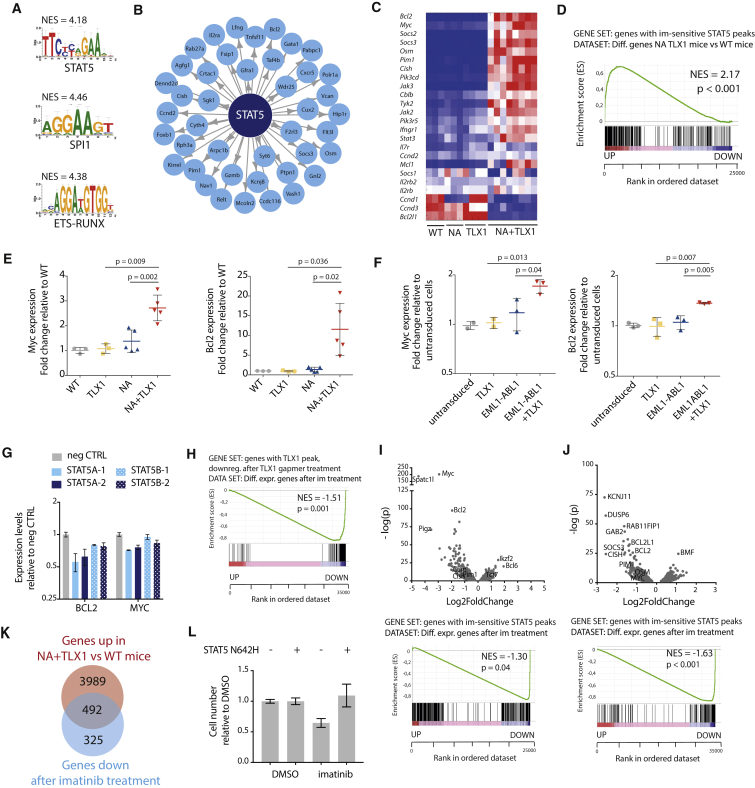
(A) Transcription factor binding motifs of the top transcription factors identified by i-CisTarget to regulate NA + TLX1 gene expression patterns. NES, normalized enrichment score.
(B) i-CisTarget transcriptional network showing genes regulated by STAT5.
(C) Heatmap representing the differential gene expression of canonical JAK-STAT signaling pathway genes in WT, NA, TLX1, and NA + TLX1 mice.
(D) Gene set enrichment analysis (GSEA) showing enrichment of STAT5 target genes (as defined by ChIP-seq) in the differentially expressed genes in NA + TLX1 mice compared with WT. NES, normalized enrichment score.
(E) qRT-PCR analysis of Myc and Bcl2 mRNA in the different transgenic mouse models. Statistical significance calculated using unpaired two-tailed t test with equal variance. Data are presented as mean ± SD.
(F) qRT-PCR analysis of Myc and Bcl2 in ex vivo immature pro T cells expressing EML1-ABL1, TLX1 or both. Statistical significance calculated using unpaired two-tailed t test with equal variance. Data are presented as mean ± SD.
(G) qRT-PCR in ALL-SIL cells after a 2-day antisense oligo-mediated knockdown of STAT5A or STAT5B. Data are presented as mean ± SD.
(H) GSEA to show enrichment of TLX1 target genes (genes with a TLX1 ChIP peak and downregulated after TLX1 gapmer treatment) in differentially expressed genes after imatinib treatment. NES, normalized enrichment score.
(I–K) Volcano plot (I) showing up- and downregulated genes (top) and GSEA to show enrichment of STAT5 target genes in differentially expressed genes (bottom) after imatinib treatment (500 nM imatinib or DMSO for 3 hr) in leukemic cells harvested from NA + TLX1 mice (n = 3; experiment was performed with cells harvested from three separate mice). (J) Volcano plot showing up- and downregulated genes (top) and GSEA to show enrichment of STAT5 target genes in differentially expressed genes (bottom) after imatinib treatment (500 nM imatinib or DMSO for 3 hr) in ALL-SIL cells (n = 3; experiment was performed as three independent repeats). (K) Venn diagram showing overlap between the upregulated genes in NA + TLX1 versus WT mice and genes that are downregulated after imatinib treatment (p = 8.7 × 10−159).
(L) Cell number of NA + TLX1 spleen cells expressing STAT5 N642H or empty vector, treated for 48 hr with 500 nM imatinib or DMSO. Data are presented as mean ± SD.
See also Figure S5.