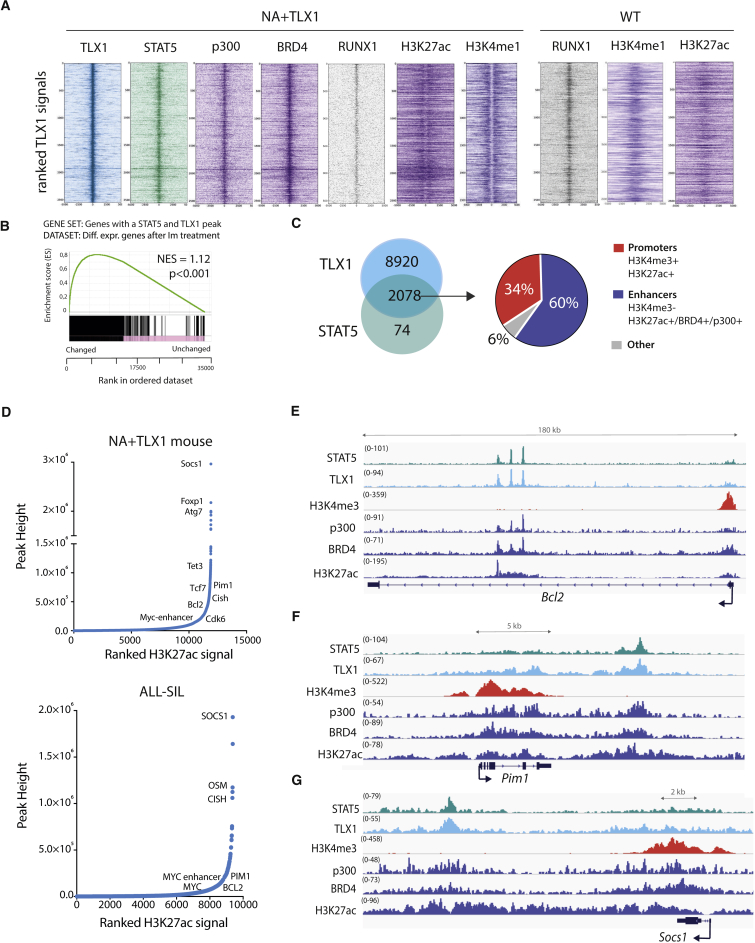
Figure 3.
STAT5 and TLX1 Co-bind Regulatory Regions throughout the Genome
(A) Centered read density heatmaps of ChIPmentation ChIP-seq signals for the binding of TLX1, STAT5, p300, BRD4, ETS1, RUNX1, and the histone marks H3K27ac and H3K4me1. Heatmaps centered and ranked on TLX1 signal strength in leukemic NA + TLX1 and WT mouse cells.
(B) GSEA comparing genes bound by both STAT5 and TLX1 and differentially expressed genes after treatment with imatinib in ALL-SIL cells.
(C) Venn diagram showing the total amount of TLX1 and STAT5 peaks that fall within the 90,804 H3K27ac peaks for NA + TLX1 leukemic cells (left). Pie chart (right) showing ChIP-seq peak co-occurrence of STAT5 and TLX1 in mouse NA + TLX1 leukemic cells in relation to promoter regions (H3K4me3+/H3K27ac+) and enhancer regions (H3K27ac+/BRD4+/p300+/H3K4me3−).
(D) Enhancer regions ranked on H3K27ac signal for NA + TLX1 leukemic cells and ALL-SIL. Only H3K27ac clusters with overlapping TLX1 peaks are shown. Gene labels are distance-based if not intragenic.
(E–G) Representative ChIP-seq tracks for canonical STAT5 target genes Bcl2 (E), Pim1 (F), and Socs1 (G) showing STAT5, TLX1, H3K4me3, p300, BRD4, and H3K27ac binding in NA + TLX1 leukemic mouse cells.