Figure 4.
TLX1 and STAT5 Bind in Newly Accessible Enhancer Regions
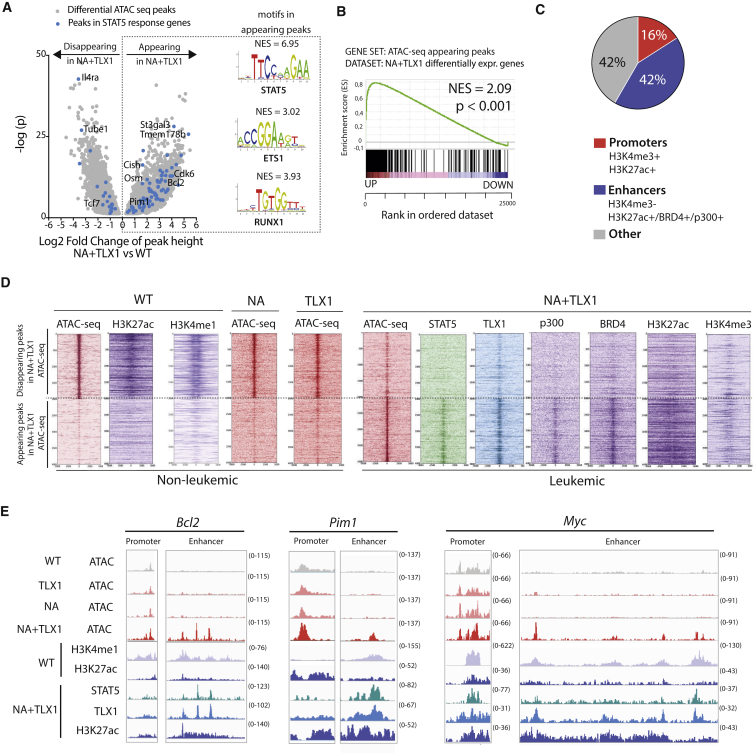
(A) Volcano plot of differential ATAC-seq peak height in CD4+CD8+ NA + TLX1 leukemic cells versus CD4+CD8+ WT cells. Peaks in STAT5 response genes are shown as blue dots. Transcription factor binding motifs enriched through i-CisTarget analysis in the appearing peaks are shown on the right. NES, normalized enrichment score.
(B) GSEA of appearing ATAC peaks in relation to differentially expressed genes in CD4+CD8+ NA + TLX1 leukemic cells versus CD4+CD8+ WT cells.
(C) Pie chart representing the genomic locations of appearing ATAC-seq peaks in CD4+CD8+ NA + TLX1 cells versus CD4+CD8+ WT cells. A promoter region is defined as a region with combined H3K4me3 and H3K27ac marks. An enhancer region is defined as a region with H3K27ac signal or BRD4 binding or p300 binding, but no H3K4me3.
(D) Read density heatmaps of ATAC-seq and ChIP-seq signals (STAT5, TLX1, p300, BRD4, H3K27ac, H3K4me1, H3K4me3) in CD4+CD8+ WT, NA, TLX1, and NA + TLX1 cells, centered around the top 2,000 appearing and disappearing ATAC-seq peaks.
(E) ATAC-seq tracks (performed in CD4+CD8+ WT, NA, TLX1, and NA + TLX1 cells) and ChIP-seq tracks (H3K4me1, H3K27ac in WT cells and STAT5, TLX1, H3K27ac in NA + TLX1 cells) at the Bcl2 locus, the Pim1 locus, and the Myc locus.
See also Figure S6.