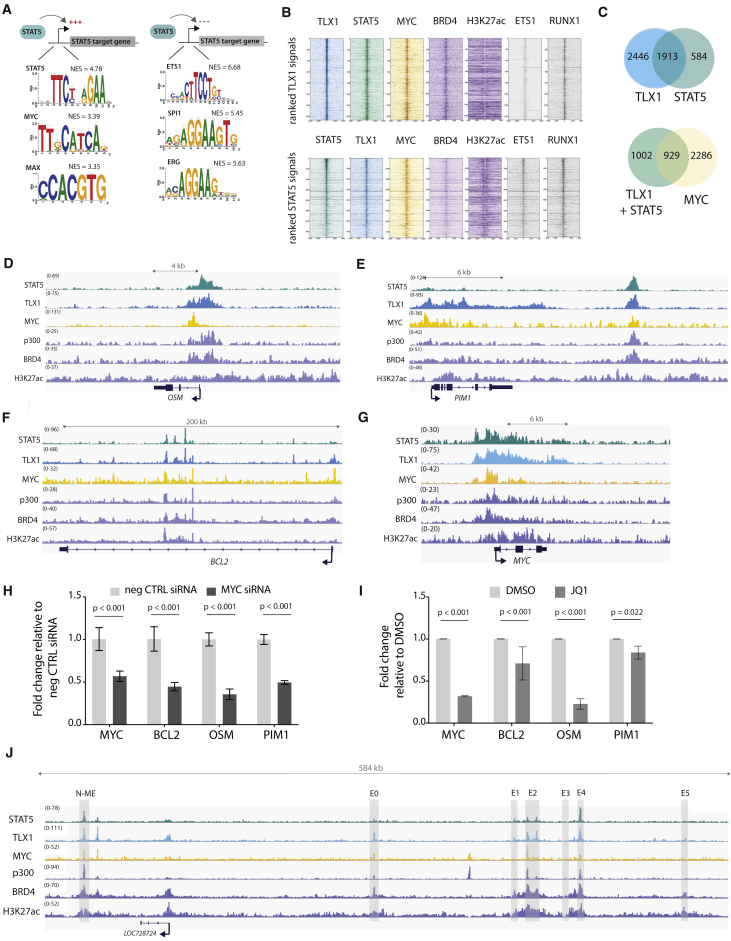
Figure 5.
MYC Co-binds and Co-regulates STAT5 and TLX1 Target Genes
(A) In silico i-CisTarget analysis for enriched transcription factor motifs found within in regulatory regions of genes that are positively or negatively regulated by STAT5.
(B) Read density heatmaps of ChIP-seq signals on TLX1 binding locations for different transcription factors or epigenetic marks ranked on TLX1 signal strength (top) or STAT5 signal strength (bottom) in ALL-SIL cells.
(C) Venn diagram showing the total amount of TLX1 and STAT5 peaks that fall within the 41,229 H3K27ac peaks (left) and the total amount of STAT5 + TLX1 peaks that overlap with MYC peaks (right) in ALL-SIL cells.
(D–G) ChIP-seq tracks (performed on ALL-SIL cells) of STAT5, TLX1, MYC, p300, BRD4, and H3K27ac at canonical STAT5 regulated genes OSM (D), PIM1 (E), BCL2 (F), and MYC (G) loci.
(H) qRT-PCR in ALL-SIL cells treated with MYC siRNAs for 48 hr. Data are presented as mean ± SD. Statistical significance calculated using unpaired two-tailed t test with equal variance.
(I) qRT-PCR of STAT5 target genes in ALL-SIL cells treated with 500 nM JQ1 for 6 hr. Data are presented as mean ± SD. Statistical significance calculated using unpaired two-tailed t test with equal variance.
(J) ChIP-seq tracks (performed on ALL-SIL cells) of STAT5, TLX1, MYC, p300, BRD4, and H3K27ac at the MYC enhancer locus, 1.4 Mb downstream of the MYC gene.