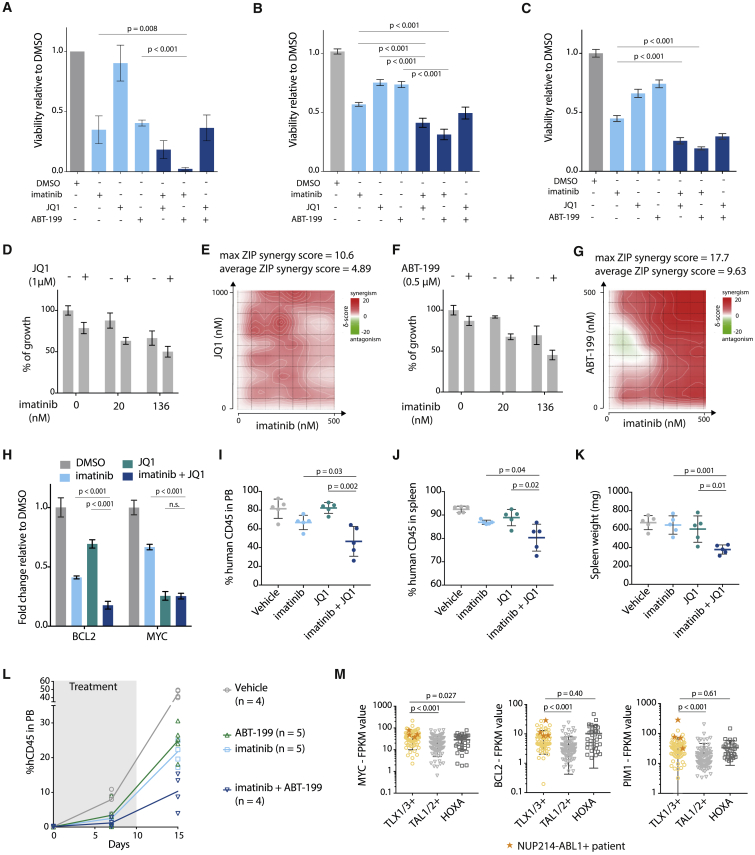
Figure 6.
Downstream Effectors of NUP214-ABL1 and TLX1 Can Be Targeted to Improve Treatment Strategies
(A) Viability of ALL-SIL cells after 48 hr treatment with 500 nM imatinib, 500 nM JQ1, 500 nM ABT-199, or a combination of these inhibitors (500 nM + 500 nM). (n = 3; experiment was performed as three independent repeats, and data are presented as mean ± SD).
(B) Viability of NUP214-ABL1+TLX3+ X12 PDX cells after 48 hr treatment with 500 nM imatinib, 500 nM JQ1, 500 nM ABT-199, or a combination of these inhibitors (500 nM + 500 nM). Statistical significance calculated using unpaired two-tailed t test with equal variance (n = 2; the experiment was performed using cells harvested from two different xenograft mice, and data are presented as mean ± SD). Statistical significance calculated using unpaired two-tailed t test with equal variance.
(C) Viability of NUP214-ABL1+TLX3+ XD82 PDX cells after 48 hr treatment with 500 nM imatinib, 500 nM JQ1, 500 nM ABT-199, or a combination of these inhibitors (500 nM + 500 nM) (n = 2; the experiment was performed using cells harvested from two different xenograft mice, and data are presented as mean ± SD). Statistical significance calculated using unpaired two-tailed t test with equal variance.
(D) Growth of X12 PDX cells after 48 hr treatment with imatinib with or without JQ1 (1 μM). Data are presented as mean ± SD.
(E) Synergy matrix plot showing δ-scores for X12 PDX cells treated with imatinib + JQ1 (average ZIP synergy score = the average δ-score for the whole range of concentrations shown in the synergy matrix; max ZIP synergy score = maximal score for a specific dose combination).
(F) Growth of X12 PDX cells after 48 hr treatment with imatinib with or without ABT-199 (0.5 μM). Data are presented as mean ± SD.
(G) Synergy matrix plot showing δ-scores for X12 PDX cells treated with imatinib + ABT-199 (average ZIP synergy score = the average δ-score for the whole range of concentrations shown in the synergy matrix; max ZIP synergy score = maximal score for a specific dose combination).
(H) qRT-PCR analysis of BCL2 and MYC mRNA expression levels in ALL-SIL cells after 6 hr of treatment with 500 nM imatinib, 500 nM JQ1, or in combination. Data are presented as mean ± SD. Statistical significance calculated using unpaired two-tailed t test with equal variance.
(I and J) Percentage of human CD45 cells detected by flow cytometry in peripheral blood samples (I) or spleen samples (J) of mice treated for 10 days with JQ1 (50 mg/kg/day), imatinib (100 mg/kg/day), or a combination of imatinib + JQ1. Data are presented as mean ± SD.
(K) Spleen weight of mice treated with JQ1, imatinib, or a combination of imatinib + JQ1. Data are presented as mean ± SD.
(L) Percentage of human CD45 detected by flow cytometry in peripheral blood samples of mice treated with ABT-199 (20 mg/kg/day), imatinib (100 mg/kg/day), or a combination of imatinib + ABT-199. Gray bar indicates treatment period. imatinib versus imatinib + ABT-199: p = 0.0048 (unpaired t test). ABT-199 versus imatinib + ABT-199: p = 0.0027 (unpaired t test). Data are presented as mean ± SD.
(M) Expression levels of MYC (left), BCL2 (middle), and PIM1 (right) in patients from different T-ALL subgroups. Patients harboring the NUP214-ABL1 fusion are represented by an orange star. Statistical significance was calculated using a Mann-Whitney test. Data are presented as mean ± SD.
See also Figure S7.