Abstract
Purpose
To map the cerebral metabolic rate of oxygen (CMRO2) by estimating the oxygen extraction fraction (OEF) from gradient echo imaging (GRE) using phase and magnitude of the GRE data.
Theory and Methods
3D multi-echo gradient echo imaging and perfusion imaging with arterial spin labeling were performed in 11 healthy subjects. CMRO2 and OEF maps were reconstructed by joint quantitative susceptibility mapping (QSM) to process GRE phases and quantitative Blood Oxygen Level Dependent (qBOLD) modeling to process GRE magnitudes. Comparisons with QSM and qBOLD alone were performed using ROI analysis, paired t-tests, and Bland-Altman plot.
Results
The average CMRO2 value in cortical gray matter across subjects were 140.4 ± 14.9, 134.1 ± 12.5, 184.6 ± 17.9 µmol/100g/min, with corresponding OEFs of 30.9 ± 3.4%, 30.0 ± 1.8%, 40.9 ± 2.4% for methods based on QSM, qBOLD, and QSM+qBOLD, respectively. QSM+qBOLD provided the highest CMRO2 contrast between gray and white matter, more uniform OEF than QSM, and less noisy OEF than qBOLD.
Conclusion
Quantitative CMRO2 mapping that fits the entire complex GRE data is feasible by combining QSM analysis of phase and qBOLD analysis of magnitude.
Keywords: Cerebral Metabolic Rate of Oxygen, Quantitative Susceptibility Mapping, quantitative Blood Oxygenation Level Dependent Imaging
INTRODUCTION
Cerebral metabolic rate of oxygen (CMRO2) and oxygen extraction fraction (OEF) maps are valuable for evaluating neurologic disorders such as ischemic stroke (1,2). Gradient echo (GRE) MR signal is highly sensitive to the strongly paramagnetic deoxyhemoglobin converted from weakly diamagnetic oxyhemoglobin. Several techniques have been proposed to estimate these maps based on quantitative modeling of either MRI magnitude or phase data. Magnitude modeling methods include quantitative imaging of extraction of oxygen and tissue consumption (QUIXOTIC) (3), calibrated fMRI (4–7) and quantitative BOLD (qBOLD) (8,9). Phase modeling methods have been used for whole brain CMRO2 measurements (10,11). Building on this, quantitative susceptibility mapping (QSM) (12,13) enables generating CMRO2 maps (14,15). The QSM-based CMRO2 methods separate voxel-wise susceptibility into non-blood tissue susceptibility (χnb) and blood susceptibility mainly driven by deoxyhemoglobin in the venous blood. The latter is then used to compute tissue oxygen consumption.
There are several challenges to the current CMRO2 mapping methods. qBOLD approximates χnb from fully oxygenated blood (9,16), but it ignores tissue iron stored in ferritin or myelin that contributes to the rich χnb contrast in the brain. QSM-based methods assume a fixed and empirical linear relationship between the venous blood volume fraction (v) and cerebral blood flow (CBF) to obtain v (17), but this relationship may vary with tissues and diseases. While qBOLD estimates v directly from the data, it assumes that χnb is constant. On the contrary, QSM-based methods assume v from CBF, whereas χnb is estimated from the data. Since both methods use the same underlying gradient echo data, we propose to combine QSM and qBOLD (QSM+qBOLD) to map the OEF and CMRO2 overcoming these assumptions.
THEORY
CMRO2 (µmol/100g/min) and OEF (%) can be expressed as
| [1] |
| [2] |
where CBF is the cerebral blood flow (ml/100g/min), [H]a is the oxygenated heme molar concentration in the arteriole (7.377 µmol/ml) estimated from [H]a = [H] · Ya, where [H] = 7.53 µmol/ml is the heme molar concentration in tissue blood assuming a hematocrit of Hct = 0.357 (18). Ya and Y are the arterial (assumed to be 0.98) and venous oxygenation. Hct, Y, and Ya are dimensionless fractions.
QSM-based CMRO2 methods require estimating two unknowns per voxel (14,15,18): venous oxygenation Y and non-blood susceptibility χnb. The venous blood volume fraction (v) is assumed to be known by an empirically derived linear relationship between cerebral blood flow (CBF) and v obtained using positron emission tomography (PET) in healthy subjects (17). However, since this assumption may break down in disease such as stroke (19), v should be treated as a parameter estimated from data. The qBOLD-based CMRO2 method (20) expresses the magnitude of the GRE signal of each voxel as a function of four unknowns: signal intensity at TE=0 (S0), transverse relaxation rate (R2), v, and Y (8,16,20). qBOLD further treats χnb, which varies spatially due to iron and myelin content, as the same as that of fully oxygenated blood. In the proposed QSM+qBOLD method, both v and χnb are treated as unknowns, and the QSM and qBOLD signal equations are combined according to maximum likelihood (ML) under Gaussian noise approximation (21). The ML estimation finds the parameter set (Y, v, χnb, S0, R2) which maximizes the probability of observing the voxel-wise susceptibility QSM and the magnitude signals S(TM) assuming respective signal models FQSM(Y, v, χnb) and FqBOLD(Y, v, χnb, S0, R2, TE). These signal models build upon prior QSM (18) and qBOLD models (20) but are modified to properly model v and χnb, as detailed below in Eqs. 4 and 7. Assuming Gaussian noise, this is equivalent to minimizing the cost function:
| [3] |
where ‖·‖2 is the L2 norm and w the weight on the QSM model of phase data. The first term in Eq. 3 is the qBOLD model of magnitude data (20):
| [4] |
where G is the macroscopic field inhomogeneity contributing to the GRE signal (see Appendix) and FBOLD is the GRE signal decay due to deoxygenated blood in vessel network: . The function fs is the signal decay by the blood vessel network (20,22), whose asymptotic behavior is for TE ≪ 1/δω, and fs(δω · TE) ≈ δω · TE for , where δω is the characteristic frequency due to the susceptibility difference between deoxygenated blood and the surrounding tissue:
| [5] |
where γ is the gyromagnetic ratio (267.513 MHz/T), B0 is the main magnetic field (3T in our study), Hct is hematocrit (0.357) (18), Δχ0 is the susceptibility difference between fully oxygenated and fully deoxygenated red blood cell (4π × 0.27ppm) (23), χba is the susceptibility of fully oxygenated blood, −108.3 ppb estimated with using Hct 0.357 (15). Note that the qBOLD assumption of neglecting susceptibility differences between non-blood tissue and fully oxygenated blood (χba = χnb) (8,16,20,24) is not made here.
The second term in Eq. 3 contains the signal equation for the susceptibility of a voxel, FQSM, which is the sum of three terms: the non-blood tissue susceptibility, the plasma susceptibility and the hemoglobin susceptibility (15,18):
| [6] |
where CBV is total blood volume (dimensionless fraction), ψHb the hemoglobin volume fraction (dimensionless) which was set to 0.0909 for tissue based on Hct 0.357 (18,25–27), χp the blood plasma susceptibility, set to −37.7 ppb (28), χoHb the oxyhemoglobin susceptibility (−813 ppb) (18,26,29), [dH]v and [dH]a the concentration (µmol/ml) of deoxyhemoglobin in venules and arterioles, respectively. [dH]v and [dH]a will be expressed in terms of Y and Ya below. Finally, ΔχHb is the susceptibility difference between deoxy- and oxy-hemoglobin (12522 ppb) (14,28).
Eq. 6 is then expressed in terms of 1) the ratio between the venous and total blood volume α = v/CBV, assumed to be constant (0.77) (30), 2) the fully oxygenated blood susceptibility, χba = ψHb · χoHb + (1 − ψHb) · χp (15), 3) the arterial oxygenation , and 4) the venous oxygenation, . This results in:
| [7] |
where the two terms represent the contribution of blood and non-blood tissue to the total susceptibility, respectively. Note that v, which was estimated from CBF in QSM-based CMRO2 mapping methods (14,15,18), is now an unknown to be determined by data.
This QSM+qBOLD model is compared with two previous methods: 1) a QSM method with minimum local variance (MLV) (18) (“QSM” hereafter), and 2) qBOLD that models the complex multi-echo gradient echo data as (20) (“qBOLD” hereafter):
| [8] |
METHODS
Data Acquisition
This study was approved by the local Institutional Review Board. Healthy volunteers were recruited (n=11; 10 males, 1 female, mean age 34 ± 12 years) for brain MRI on a 3T scanner (HDxt, GE Healthcare) using an 8-channel brain receiver coil. After obtaining consent, all subjects were instructed to avoid caffeine or alcohol intake 24 hours prior to the MRI.
MRI was performed in the resting state (15) using a 3D fast spin echo (FSE) arterial spin labeling (ASL) sequence (31–33), a 3D multi-echo spoiled gradient echo (GRE) sequence (13,34,35), and an inversion prepared T1w SPGR sequence (BRAVO) (36). The 3D FSE ASL sequence parameters were: 20 cm FOV, 1.56 mm in-plane resolution, 3.5 mm slice thickness, 1500 ms labeling period, 1525 ms post-label delay, 976.6 Hz/pixel bandwidth, spiral sampling of 8 interleaves with 512 readout points per leaf, 35 axial slices, 10.1 ms TE, 4533 ms TR, and 3 signal averages. The 3D GRE sequence parameters were: 0.78 mm in-plane resolution, 1.2 mm slice thickness, volume coverage identical to the 3D FSE ASL sequence, 7 equally spaced echoes, 2.3 ms for the first TE, 3.9 ms echo spacing, 30.5 ms TR, 488.3 Hz/pixel band width, and 15o flip angle. The pulse sequence was flow-compensated in all three directions (34). The inversion prepared T1w SPGR sequence parameters were: 0.78 mm in-plane resolution, 1.2 mm slice thickness, volume coverage identical to the 3D FSE ASL sequence, 2.92 ms TE, 7.69 ms TR, 450 ms prep time, 195.2 Hz/pixel bandwidth, and 15o flip angle.
Image Processing
QSM reconstruction was performed as follows: first, an adaptive quadratic-fit of the GRE phase was used to estimate the total field (34). Second, the Projection onto dipole fields (PDF) method was used to obtain the local field (35). Finally, the Morphology Enabled Dipole Inversion (MEDI) algorithm was used to compute susceptibility (12,13,37). The susceptibility values were referenced to the susceptibility of cerebrospinal fluid (CSF) averaged over a manually drawn ROI on the first echo of the GRE acquisition. CBF maps (ml/100g/min) were generated from the ASL data using the FuncTool software package (GE Healthcare, Waukesha, WI, USA). All images were co-registered and interpolated to the resolution of the QSM maps using the FSL FLIRT algorithm (38,39).
Optimization
To improve convergence behavior during nonlinear fitting, the five unknowns v, Y, χnb, S0, R2 were scaled to have roughly the same order of magnitude as the initial guess: , where x is the unknown in the original scale and x0 is the corresponding initial guess. Initial guesses were obtained as follows: Y0 was estimated from the straight sinus (SS) as the global constraint in previous QSM-based CMRO2 studies (15,18), Y0 = Ya · (1 − OEFss/Hctratio where Hctratio is the hematocrit ratio (0.759) between large vessels and brain tissue (25) and OEFss = 1− Yss/Ya (Yss was estimated from QSM in SS with setting ψHb = 0.1197 for large veins (18,26,27), v = 1 and χnb = 0 in Eq. 7). v0 was set using the linear relationship between CBF and v (14,15,18). χnb,0 was set as the susceptibility of fullyoxygenated blood as in the qBOLD methods (8,16,20). and R2,0 were obtained by performing a mono-exponential fit against Eq. 4 after fixing the values of G (40) and FBOLD with the initial values Y0, v0, and χnb,0. The function G was calculated using the voxel spread function (VSF) method (41). For some voxels the initials guess for Y was not appropriate leading to an increase in signal as a function of TE after dividing out the FBOLD contribution. This then led to a negative R2,0. To avoid this situation, Y was gradually increased to insure a positive R2,0.
To speed up the FBOLD calculation, we used a Taylor expansion of the hypergeometric function (1F2) up to the 60th order (20). The lower and upper bounds were set to 0.5 and 2 for each scaled unknown, except for Y, which used the 0.1 and 0.9 bounds before scaling. At the start of each optimization, both the QSM and qBOLD terms in Eq. 3 were normalized by their respective values evaluated with the initial guess to compensate for the arbitrary scale of input MRI data. The tuning parameter w was selected using the L-curve method (42). Corners of the L-curves for two randomly chosen subjects were both located at w = 100 (Fig. 1). The limited-memory Broyden-Fletcher-Goldfarb-Shanno-Bound algorithm was used for the constrained optimization (43,44). The optimization was stopped when the relative residual , with Ei as the energy of the ith iteration, was smaller than 0.005.
Figure 1.
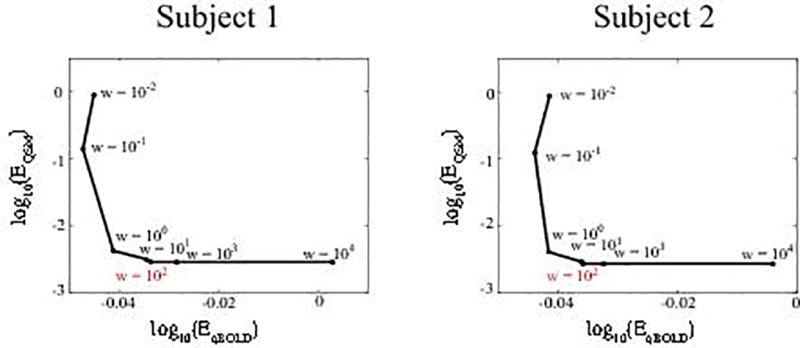
L-curves in two randomly selected subjects to determine tuning parameter w. The L-curve corners were located at w=100 in both subjects. EqBOLD and EQSM indicate the energy terms in Eq. 3.
For “QSM”, the optimization followed the previous QSM-based method using the MLV method and global CMRO2 constraint (18). For “qBOLD”, the optimization was performed voxel-wise, following (20). The parameter scaling, initial guess, and boundary conditions were set in the same manner as for QSM+qBOLD. In addition, the initial frequency shift Δf0 was set to the total field value as obtained in the QSM process (13).
Statistical Analysis
ROI analyses (mean and standard deviation), paired t-tests (at 0.01 significance level), and Bland-Altman analyses were performed to compare CMRO2 and OEF values between the QSM-based (18), qBOLD (20), and QSM+qBOLD methods. For the ROIs, the bilateral anterior (ACA), middle (MCA) and posterior (PCA) cerebral artery vascular territories (VT) in the cortical gray matter (CGM) and white matter (WM) masks were constructed based on T1-weighted images by a neuroradiologist with 10 years of experience.
RESULTS
The CMRO2 in CGM were 140.4 ± 14.9, 134.1 ± 12.5, 184.6 ± 17.9 µmol/100g/min (N=11) for the QSM, qBOLD, and QSM+qBOLD models, respectively. These corresponded to OEFs of 30.9 ± 3.4%, 30.0 ± 1.8%, 40.9 ± 2.2%, respectively (Supporting Table S1).
In Fig. 2, compared to QSM and qBOLD, QSM+qBOLD showed higher CMRO2 contrast between CGM and WM: 21.9 ± 9.4 (p<0.01), 36.0 ± 6.4 (p<0.01), 54.6 ± 10.3 µmol/100g/min (N=11) for the QSM, qBOLD, and QSM+qBOLD models, respectively. QSM showed a smoother CMRO2 map than QSM+qBOLD, except for a sharp transition at the GM/WM boundary.
Figure 2.
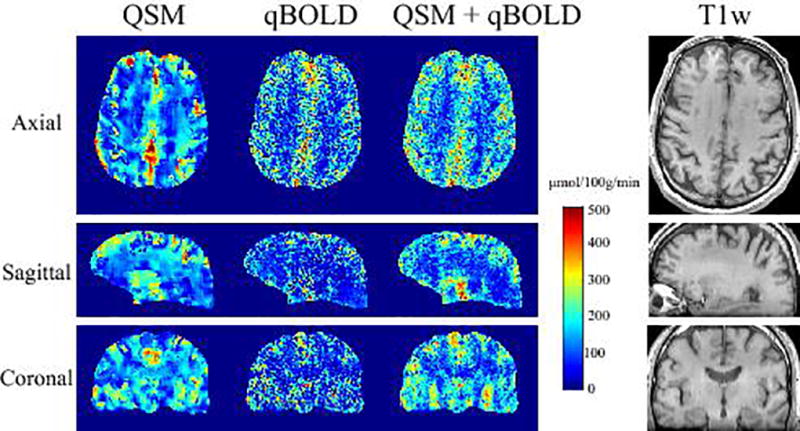
CMRO2 map in axial, sagittal, and coronal sections from a subject reconstructed using QSM, qBOLD, and QSM+qBOLD methods. The corresponding T1-weighted anatomical images are shown on the right. QSM+qBOLD shows higher GM/WM contrast than QSM or qBOLD.
In Fig. 3, QSM+qBOLD showed higher OEF than the two individual methods; the OEF values for whole brain were 34.5 ± 3.1 % (p<0.01), 29.8 ± 2.1% (p<0.01), and 39.3 ± 2.2 % (N=11) for QSM, qBOLD, and QSM+qBOLD, respectively. QSM+qBOLD showed a relatively uniform OEF map as compared to QSM, and a less noisy OEF map than qBOLD. The OEF standard deviations in CGM was lower for QSM+qBOLD than for each individual methods: 16.3 ± 0.9 % (p<0.01), 19.4 ± 1.5 % (p<0.01), and 12.3 ± 1.2 % (N=11) for QSM, qBOLD, and QSM+qBOLD, respectively. The venous blood volume v was slightly higher for QSM+qBOLD as compared to QSM and qBOLD: in CGM, the values were 4.3 ± 0.4 % (p<0.01), 4.1 ± 0.4% (p<0.01), and 4.5 ± 0.4%, and in WM, 3.5 ± 0.3 % (p=0.011) and 3.2 ± 0.3% (p<0.01), and 3.5 ± 0.3% for QSM, qBOLD and QSM+qBOLD, respectively. χnb from QSM and QSM+qBOLD showed similar contrast at the CGM/WM boundary, it was higher in CGM than in nearby WM. Average χnb values were similar between QSM and QSM+qBOLD as compared to qBOLD (−108.3 ppb assumed for the whole brain): −12.3 ± 3.7 ppb (p<0.01) and −19.8 ± 3.5 ppb in CGM, and −15.3 ± 3.9 ppb (p<0.01) and −18.7 ± 3.5 ppb in WM (N=11) for QSM and QSM+qBOLD, respectively. The v map of the QSM model and the χnb of the qBOLD model were not obtained through optimization, but given by a priori assumptions.
Figure 3.
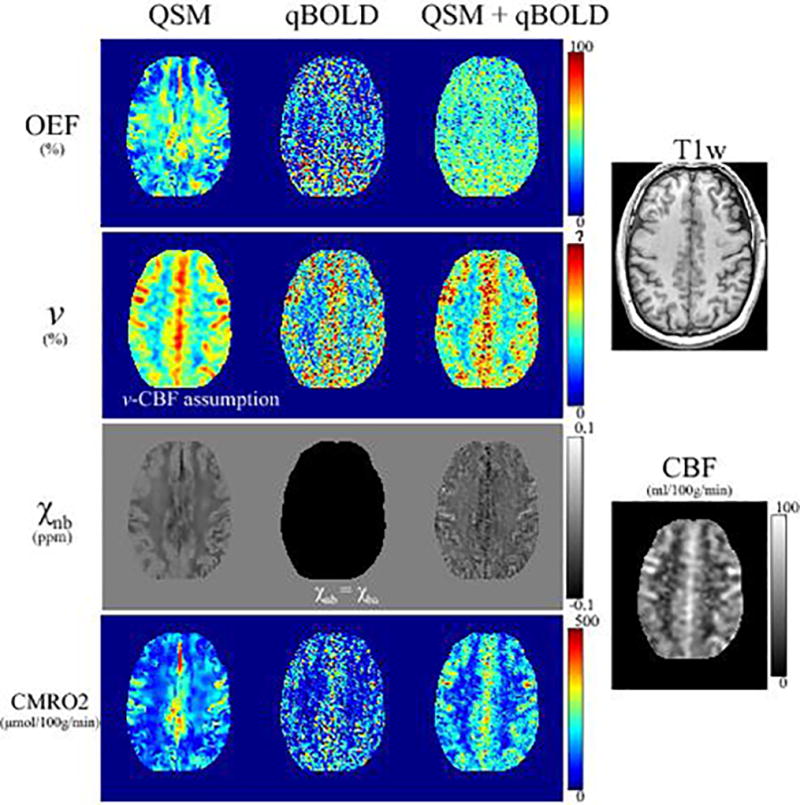
OEF, venous volume (v), non-blood tissue susceptibility (χnb), and CMRO2 maps in a second subject reconstructed using QSM, qBOLD, and QSM+qBOLD models. The OEF obtained using QSM+qBOLD is more uniform than that of QSM, and is greater than that of qBOLD.
In Fig. 4, QSM+qBOLD showed higher CMRO2 and OEF values than both QSM and qBOLD in all ACA, MCA, PCA, and WM ROIs (p<0.01). All three methods showed lower CMRO2 in WM than in ACA, MCA, and PCA (p<0.01).
Figure 4.
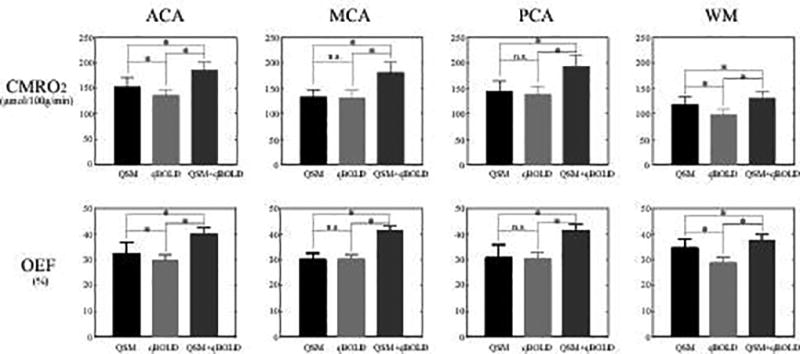
ROI comparison of CMRO2 and OEF in ACA, MCA, PCA, and WM among QSM (black), qBOLD (gray), and QSM+qBOLD (dark gray). In all ROIs, QSM+qBOLD provides higher CMRO2 and OEF values than QSM or qBOLD (p<0.01). * p < 0.01 (paired t-test). “n.s.” indicates that the difference is not significant (p>0.1).
In the Bland-Altman plots (Fig. 5), the mean differences were 43.5 µmol/100g/min (QSM vs QSM+qBOLD) and 51.2 µmol/100g/min (qBOLD vs QSM+qBOLD) for CMRO2 (p<0.01), and 9.8 % (QSM vs QSM+qBOLD) and 10.9 % (qBOLD vs QSM+qBOLD) for OEF (p<0.01).
Figure 5.
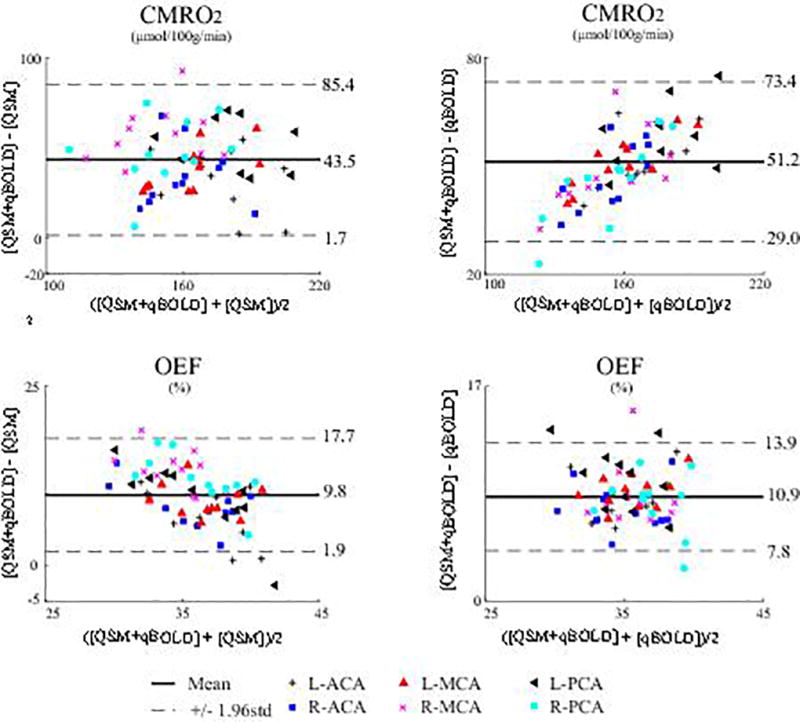
Bland-Altman plots comparing OEF and CMRO2 values in VT ROIs between QSM+qBOLD and the two reference methods, QSM and qBOLD. The mean differences are 43.5 µmol/100g/min (QSM vs QSM+qBOLD) and 51.2 µmol/100g/min (qBOLD vs QSM+qBOLD) for CMRO2 (p<0.01), and 9.8 % (QSM vs QSM+qBOLD) and 10.9 % (qBOLD vs QSM+qBOLD) for OEF (p<0.01).
DISCUSSION
Our results demonstrate the feasibility of a QSM+qBOLD method for mapping CMRO2 by comprehensive modeling of both the magnitude and phase of multi-echo gradient echo data. When either QSM modeling phase or qBOLD modeling magnitude are used individually to estimate CMRO2, a number of assumptions are required: a linear relationship between CBF and v and the minimum local variance of the estimated parameters within a tissue block for the QSM-based method (18), and constant non-blood tissue susceptibility for the qBOLD method (16,20). These assumptions may cause errors and are eliminated in the proposed QSM+qBOLD method.
Compared to QSM and qBOLD alone, the proposed QSM+qBOLD shows clearer GM/WM CMRO2 contrasts (Fig. 2), more uniform OEF compared to QSM (Fig. 3), and better agreement with results presented using other independent CMRO2 estimation methods such as PET (45). By overcoming the limited availability of PET, MRI provides greater accessibility for patient studies. Without any vascular challenge, QSM+qBOLD can be readily applied to study damage to vital organs caused by oxygen-deficiency, including Alzheimer’s disease (AD) (46,47), multiple sclerosis (48,49), and ischemia in stroke (1,50).
Compared to qBOLD, QSM+qBOLD shows higher OEF (Figs. 3, 4, and 5), with 39.3 ± 2.2 % for the whole brain; this is in a good agreement with results from previous PET studies which have OEFs of 35 ± 7 % (51), 42.6 ± 5.1 % (52), 41 ± 6 % (53), and 40 ± 9 % (54). This difference is not driven by venous blood volume (v), which was found to be similar between qBOLD and QSM+qBOLD (Fig. 3). Instead, the higher OEF may be explained by the inclusion of non-blood tissue susceptibility (χnb) as a variable, which, in qBOLD, is assumed to be equal to the susceptibility of fully oxygenated blood (χba). The resulting χnb (−19.8 ± 3.5 ppb in CGM) is greater than χba (−108.3 ppb). To obtain the same measured susceptibility difference δω in Eq. 5 when χba − χnb < 0, the venous oxygenation Y needs to decrease, which leads to a higher OEF (Eqs. 2 and 5). This suggests that inclusion of χnb was a factor in the observed OEF increase.
QSM+qBOLD shows higher OEF average than QSM for whole brain (39.3 ± 2.2 % vs 34.5 ± 3.1 %). A likely cause of this may lie in the use of the global constraint in QSM (18), which is not used in QSM+qBOLD. The global constraint from straight sinus (SS) might not precisely estimate the average oxygentation in the whole brain because SS may not represent all the draining veins (55,56) that are known to have considerable variation, e.g. 47.7% ~ 75.3% (57).
At the GM/WM boundary in the CMRO2 map, the QSM method shows a sharp transition, whereas the QSM+qBOLD method reveals a gradual change (Figs. 2 and 3). The sharp GM/WM edge in QSM may be caused by separate optimization for the GM and WM (18); in the QSM+qBOLD method, the optimization was performed for whole brain. The CMRO2 GM/WM contrast in QSM+qBOLD is primarily driven by CBF since OEF map looks fairly uniform (Fig. 3). A lack of CMRO2 GM/WM contrast in QSM is not uniform and more prominent in areas where OEF appears underestimated (Figure 3, top and bottom left). Errors in GM/WM segmentation or smoothing by the MLV assumption may have caused these local underestimations.
The CMRO2 and OEF maps are smoother using the QSM method (Figs. 2 and 3). This may be caused by the use of MLV in the QSM method (18), which assumes that CMRO2 and χnb are constant within small blocks of brain tissue, to overcome the underdetermined system (2 unknowns and 1 equation per voxel) when no challenge is used. The MLV assumption in the QSM method is highly dependent on the block setting, e.g. block size and number of block shifts. The QSM+qBOLD model eliminates this MLV assumption using qBOLD modeling of the magnitude data.
The non-blood tissue susceptibility χnb shows a clear GM/WM contrast using QSM+qBOLD (Fig. 3). This is consistent with the difference in myelin content between CGM and WM (58).
The QSM+qBOLD model may still be affected by residual limitations in QSM and qBOLD models. The qBOLD term ignores the detailed microstructure of brain tissue, including myelin water components and magnetic anisotropy of myelin in white matter. The qBOLD Eq.4 may be expanded with 2 different R2s to account for myelin water, and fiber orientation dependent effects in Eq.5 to consider myelin magnetic anisotropy. The empirical L-curve analysis was used to determine relative weighting (w) in Eq.3. Additional studies are needed to investigate the influence of the weighting w, measurement error and differences in acquisition schemes. For instance, the propagation of bias and error in the estimated susceptibility into QSM+qBOLD measurments will depend on the choice of w. Spatial resolution will play a role in the choice of w: while smaller voxels typically improve QSM estimates (59), the resulting signal to noise ratio (SNR) loss may affect qBOLD estimates negatively, since it requires high SNR (16).
While QSM+qBOLD removed the linear CBF/CBV assumption in QSM and the constant non-blood tissue susceptibility assumption in qBOLD, it retains a constant ratio between venous and total blood volume (0.77), constant tissue hematocrit (0.357) and constant oxygenated heme molar concentration in the arteriole (7.377 µmol/ml) for the whole brain, which may vary among subjects and diseases. The ASL-measured CBF is known to contain errors, particularly in WM (60) and maybe deteriorated with the usage of shorter post-label delay in this study (1525 ms as used in the clinical protocol at our institution) than the recommended value of 2000 ms (32), which will propagate to CMRO2. The OEF and v estimation in large veins might be errorous because they were treated in the same way as normal brain tissue. This error could be suppressed using the v = 1 prior for large veins. The QSM+qBOLD optimization (Eq. 3) is non-convex, with convergence susceptible to dependency on solver implementation, initial guesses, parameter scaling, and stopping criterion. A reference standard such as 15O PET is needed to validate CMRO2 methods. Finally, the application of this method and its validation for a general patient population remains to be investigated.
In conclusion, our study demonstrated the feasibility of a comprehensive quantitative CMRO2 mapping method by combining QSM and qBOLD-based methods. In healthy subjects, the CMRO2 map obtained with QSM+qBOLD shows better GM/WM contrast as compared to an individual QSM or qBOLD-based method, while the OEF map appeared more uniform compared to the QSM-based method.
Supplementary Material
Supporting Table S1. CMRO2 (µmol/100g/min) and OEF (%) for the cortical gray matter for 11 subjects (Mean ± Standard Deviation)
Acknowledgments
We thank Kelly McCabe Gillen, PhD, for her assistance in manuscript editing.
APPENDIX
In Eq. 4, the macroscopic field inhomogeneity contributing to the GRE signal (G) can be estimated via the voxel spread function (20,40,41):
where TE1 is the first echo time, bm is the average magnetic field in the voxel m, ηnm,j is the voxel spread function (the contribution of voxel m to voxel n) in jth direction (x, y, and z): ηnm,j = Σqj sinc (qj − qm,j (TE)) · e2πi·qj(n−m) where , gm,j and ϕm,j are the gradient of the background field and the first echo phase, respectively, and kj is k-space , Nj and aj are matrix size and voxel size in the jth direction, respectively.
References
- 1.Baron JC. Mapping the ischaemic penumbra with PET: implications for acute stroke treatment. Cerebrovascular diseases (Basel, Switzerland) 1999;9(4):193–201. doi: 10.1159/000015955. [DOI] [PubMed] [Google Scholar]
- 2.Gupta A, Baradaran H, Schweitzer AD, Kamel H, Pandya A, Delgado D, Wright D, Hurtado-Rua S, Wang Y, Sanelli PC. Oxygen Extraction Fraction and Stroke Risk in Patients with Carotid Stenosis or Occlusion: A Systematic Review and Meta-Analysis. American Journal of Neuroradiology. 2014;35(2):250–255. doi: 10.3174/ajnr.A3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolar DS, Rosen BR, Sorensen AG, Adalsteinsson E. QUantitative Imaging of eXtraction of oxygen and TIssue consumption (QUIXOTIC) using venular-targeted velocity-selective spin labeling. Magn Reson Med. 2011;66(6):1550–1562. doi: 10.1002/mrm.22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoge RD. Calibrated FMRI. NeuroImage. 2012;62(2):930–937. doi: 10.1016/j.neuroimage.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 5.Gauthier CJ, Hoge RD. Magnetic resonance imaging of resting OEF and CMRO(2) using a generalized calibration model for hypercapnia and hyperoxia. NeuroImage. 2012;60(2):1212–1225. doi: 10.1016/j.neuroimage.2011.12.056. [DOI] [PubMed] [Google Scholar]
- 6.Wise RG, Harris AD, Stone AJ, Murphy K. Measurement of OEF and absolute CMRO 2: MRI-based methods using interleaved and combined hypercapnia and hyperoxia. Neuroimage. 2013;83:135–147. doi: 10.1016/j.neuroimage.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulte DP, Kelly M, Germuska M, Xie J, Chappell MA, Okell TW, Bright MG, Jezzard P. Quantitative measurement of cerebral physiology using respiratory-calibrated MRI. Neuroimage. 2012;60(1):582–591. doi: 10.1016/j.neuroimage.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He X, Yablonskiy DA. Quantitative BOLD: mapping of human cerebral deoxygenated blood volume and oxygen extraction fraction: default state. Magn Reson Med. 2007;57(1):115–126. doi: 10.1002/mrm.21108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Sukstanskii AL, Yablonskiy DA. Optimization strategies for evaluation of brain hemodynamic parameters with qBOLD technique. Magnetic resonance in medicine. 2013;69(4):1034–1043. doi: 10.1002/mrm.24338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wehrli FW, Fan AP, Rodgers ZB, Englund EK, Langham MC. Susceptibility-based time-resolved whole-organ and regional tissue oximetry. NMR in biomedicine. 2017;30(4) doi: 10.1002/nbm.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan AP, Benner T, Bolar DS, Rosen BR, Adalsteinsson E. Phase-based regional oxygen metabolism (PROM) using MRI. Magn Reson Med. 2012;67(3):669–678. doi: 10.1002/mrm.23050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Rochefort L, Liu T, Kressler B, Liu J, Spincemaille P, Lebon V, Wu J, Wang Y. Quantitative susceptibility map reconstruction from MR phase data using bayesian regularization: validation and application to brain imaging. Magn Reson Med. 2010;63(1):194–206. doi: 10.1002/mrm.22187. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Liu T. Quantitative susceptibility mapping (QSM): Decoding MRI data for a tissue magnetic biomarker. Magn Reson Med. 2015;73(1):82–101. doi: 10.1002/mrm.25358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Liu T, Gupta A, Spincemaille P, Nguyen TD, Wang Y. Quantitative mapping of cerebral metabolic rate of oxygen (CMRO2) using quantitative susceptibility mapping (QSM) Magnetic Resonance in Medicine. 2015;74(4):945–952. doi: 10.1002/mrm.25463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Zhou D, Nguyen TD, Spincemaille P, Gupta A, Wang Y. Cerebral metabolic rate of oxygen (CMRO2) mapping with hyperventilation challenge using quantitative susceptibility mapping (QSM) Magnetic resonance in medicine. 2017;77(5):1762–1773. doi: 10.1002/mrm.26253. [DOI] [PubMed] [Google Scholar]
- 16.Yablonskiy DA, Sukstanskii AL, He X. BOLD-based Techniques for Quantifying Brain Hemodynamic and Metabolic Properties – Theoretical Models and Experimental Approaches. NMR in biomedicine. 2013;26(8):963–986. doi: 10.1002/nbm.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leenders KL, Perani D, Lammertsma AA, Heather JD, Buckingham P, Healy MJ, Gibbs JM, Wise RJ, Hatazawa J, Herold S, et al. Cerebral blood flow, blood volume and oxygen utilization. Normal values and effect of age. Brain : a journal of neurology. 1990;113(Pt 1):27–47. doi: 10.1093/brain/113.1.27. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Cho J, Zhou D, Nguyen TD, Spincemaille P, Gupta A, Wang Y. Quantitative susceptibility mapping-based cerebral metabolic rate of oxygen mapping with minimum local variance. Magn Reson Med. 2017 doi: 10.1002/mrm.26657. [DOI] [PubMed] [Google Scholar]
- 19.Grandin CB, Duprez TP, Smith AM, Mataigne F, Peeters A, Oppenheim C, Cosnard G. Usefulness of Magnetic Resonance–Derived Quantitative Measurements of Cerebral Blood Flow and Volume in Prediction of Infarct Growth in Hyperacute Stroke. Stroke. 2001;32(5):1147–1153. doi: 10.1161/01.str.32.5.1147. [DOI] [PubMed] [Google Scholar]
- 20.Ulrich X, Yablonskiy DA. Separation of cellular and BOLD contributions to T2* signal relaxation. Magnetic resonance in medicine. 2016;75(2):606–615. doi: 10.1002/mrm.25610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogg R, McKean J, Craig A. Introduction to mathematical statistics: Pearson Education. 2005 [Google Scholar]
- 22.Sukstanskii AL, Yablonskiy DA. Theory of FID NMR signal dephasing induced by mesoscopic magnetic field inhomogeneities in biological systems. Journal of magnetic resonance (San Diego, Calif: 1997) 2001;151(1):107–117. doi: 10.1006/jmre.2001.2363. [DOI] [PubMed] [Google Scholar]
- 23.Yablonskiy DA, Haacke EM. Theory of NMR signal behavior in magnetically inhomogeneous tissues: the static dephasing regime. Magnetic resonance in medicine. 1994;32(6):749–763. doi: 10.1002/mrm.1910320610. [DOI] [PubMed] [Google Scholar]
- 24.He X, Yablonskiy DA. Biophysical mechanisms of phase contrast in gradient echo MRI. Proceedings of the National Academy of Sciences. 2009;106(32):13558–13563. doi: 10.1073/pnas.0904899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai F, Nakazawa K, Tazaki Y, Ishii K, Hino H, Igarashi H, Kanda T. Regional cerebral blood volume and hematocrit measured in normal human volunteers by single-photon emission computed tomography. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1985;5(2):207–213. doi: 10.1038/jcbfm.1985.27. [DOI] [PubMed] [Google Scholar]
- 26.Savicki JP, Lang G, Ikeda-Saito M. Magnetic susceptibility of oxy- and carbonmonoxyhemoglobins. Proceedings of the National Academy of Sciences. 1984;81(17):5417–5419. doi: 10.1073/pnas.81.17.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffman R. Hematology: Basic Principles and Practice: Churchill Livingstone. 2005 [Google Scholar]
- 28.Spees WM, Yablonskiy DA, Oswood MC, Ackerman JJ. Water proton MR properties of human blood at 1.5 Tesla: magnetic susceptibility, T(1), T(2), T*(2), and non-Lorentzian signal behavior. Magn Reson Med. 2001;45(4):533–542. doi: 10.1002/mrm.1072. [DOI] [PubMed] [Google Scholar]
- 29.Cerdonio M, Morante S, Torresani D, Vitale S, DeYoung A, Noble RW. Reexamination of the evidence for paramagnetism in oxy- and carbonmonoxyhemoglobins. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(1):102–103. doi: 10.1073/pnas.82.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An H, Lin W. Cerebral venous and arterial blood volumes can be estimated separately in humans using magnetic resonance imaging. Magn Reson Med. 2002;48(4):583–588. doi: 10.1002/mrm.10257. [DOI] [PubMed] [Google Scholar]
- 31.Dai W, Garcia D, de Bazelaire C, Alsop DC. Continuous flow-driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn Reson Med. 2008;60(6):1488–1497. doi: 10.1002/mrm.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alsop DC, Detre JA, Golay X, Gunther M, Hendrikse J, Hernandez-Garcia L, Lu H, MacIntosh BJ, Parkes LM, Smits M, van Osch MJ, Wang DJ, Wong EC, Zaharchuk G. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med. 2015;73(1):102–116. doi: 10.1002/mrm.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buxton RB, Frank LR, Wong EC, Siewert B, Warach S, Edelman RR. A general kinetic model for quantitative perfusion imaging with arterial spin labeling. Magnetic resonance in medicine. 1998;40(3):383–396. doi: 10.1002/mrm.1910400308. [DOI] [PubMed] [Google Scholar]
- 34.Xu B, Liu T, Spincemaille P, Prince M, Wang Y. Flow compensated quantitative susceptibility mapping for venous oxygenation imaging. Magn Reson Med. 2014;72(2):438–445. doi: 10.1002/mrm.24937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu T, Khalidov I, de Rochefort L, Spincemaille P, Liu J, Tsiouris AJ, Wang Y. A novel background field removal method for MRI using projection onto dipole fields (PDF) NMR in biomedicine. 2011;24(9):1129–1136. doi: 10.1002/nbm.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mugler JP, Brookeman JR., 3rd Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE) Magn Reson Med. 1990;15(1):152–157. doi: 10.1002/mrm.1910150117. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Liu T, de Rochefort L, Ledoux J, Khalidov I, Chen W, Tsiouris AJ, Wisnieff C, Spincemaille P, Prince MR, Wang Y. Morphology enabled dipole inversion for quantitative susceptibility mapping using structural consistency between the magnitude image and the susceptibility map. NeuroImage. 2012;59(3):2560–2568. doi: 10.1016/j.neuroimage.2011.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical image analysis. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 39.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 40.Yablonskiy DA, Sukstanskii AL, Luo J, Wang X. Voxel spread function method for correction of magnetic field inhomogeneity effects in quantitative gradient-echo-based MRI. Magnetic resonance in medicine. 2013;70(5):1283–1292. doi: 10.1002/mrm.24585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulrich X, Yablonskiy DA. Enhancing image contrast in human brain by voxel spread function method; In Proceedings of the 22nd Annual Meeting of ISMRM; Milan, Italy. Abstract 3197. [Google Scholar]
- 42.Hansen PC. Analysis of discrete ill-posed problems by means of the L-curve. SIAM review. 1992;34(4):561–580. [Google Scholar]
- 43.Liu DC, Nocedal J. On the limited memory BFGS method for large scale optimization. Mathematical programming. 1989;45(1):503–528. [Google Scholar]
- 44.Byrd RH, Lu P, Nocedal J, Zhu C. A limited memory algorithm for bound constrained optimization. SIAM Journal on Scientific Computing. 1995;16(5):1190–1208. [Google Scholar]
- 45.Mintun MA, Raichle ME, Martin WR, Herscovitch P. Brain oxygen utilization measured with O-15 radiotracers and positron emission tomography. J Nucl Med. 1984;25(2):177–187. [PubMed] [Google Scholar]
- 46.Sun X, He G, Qing H, Zhou W, Dobie F, Cai F, Staufenbiel M, Huang LE, Song W. Hypoxia facilitates Alzheimer's disease pathogenesis by up-regulating BACE1 gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(49):18727–18732. doi: 10.1073/pnas.0606298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Acosta-Cabronero J, Williams GB, Cardenas-Blanco A, Arnold RJ, Lupson V, Nestor PJ. In vivo quantitative susceptibility mapping (QSM) in Alzheimer's disease. PloS one. 2013;8(11):e81093. doi: 10.1371/journal.pone.0081093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapp BD, Stys PK. Virtual hypoxia and chronic necrosis of demyelinated axons in multiple sclerosis. The Lancet Neurology. 2009;8(3):280–291. doi: 10.1016/S1474-4422(09)70043-2. [DOI] [PubMed] [Google Scholar]
- 49.Wisnieff C, Ramanan S, Olesik J, Gauthier S, Wang Y, Pitt D. Quantitative susceptibility mapping (QSM) of white matter multiple sclerosis lesions: Interpreting positive susceptibility and the presence of iron. Magn Reson Med. 2015;74(2):564–570. doi: 10.1002/mrm.25420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kudo K, Liu T, Murakami T, Goodwin J, Uwano I, Yamashita F, Higuchi S, Wang Y, Ogasawara K, Ogawa A, Sasaki M. Oxygen extraction fraction measurement using quantitative susceptibility mapping: Comparison with positron emission tomography. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2016;36(8):1424–1433. doi: 10.1177/0271678X15606713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carpenter DA, Grubb RL, Jr, Tempel LW, Powers WJ. Cerebral oxygen metabolism after aneurysmal subarachnoid hemorrhage. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1991;11(5):837–844. doi: 10.1038/jcbfm.1991.143. [DOI] [PubMed] [Google Scholar]
- 52.Yamauchi H, Fukuyama H, Nagahama Y, Nabatame H. Significance of increased oxygen extraction fraction in five-year prognosis of major cerebral arterial occlusive diseases. The Journal of Nuclear Medicine. 1999;40(12):1992. [PubMed] [Google Scholar]
- 53.Diringer Michael N, Yundt Kent, Videen Tom O, Adams Robert E, Zazulia Allyson R, Deibert Ellen, Aiyagari Venkatesh, Dacey J Ralph G, Grubb J Robert L, Powers William J. No reduction in cerebral metabolism as a result of early moderate hyperventilation following severe traumatic brain injury. Journal of neurosurgery. 2000;92(1):7–13. doi: 10.3171/jns.2000.92.1.0007. [DOI] [PubMed] [Google Scholar]
- 54.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proceedings of the National Academy of Sciences. 2001;98(2):676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doepp F, Schreiber SJ, von Munster T, Rademacher J, Klingebiel R, Valdueza JM. How does the blood leave the brain? A systematic ultrasound analysis of cerebral venous drainage patterns. Neuroradiology. 2004;46(7):565–570. doi: 10.1007/s00234-004-1213-3. [DOI] [PubMed] [Google Scholar]
- 56.Meder JF, Chiras J, Roland J, Guinet P, Bracard S, Bargy F. Venous territories of the brain. Journal of neuroradiology Journal de neuroradiologie. 1994;21(2):118–133. [PubMed] [Google Scholar]
- 57.Fan AP, Bilgic B, Gagnon L, Witzel T, Bhat H, Rosen BR, Adalsteinsson E. Quantitative oxygenation venography from MRI phase. Magnetic resonance in medicine. 2014;72(1):149–159. doi: 10.1002/mrm.24918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langkammer C, Krebs N, Goessler W, Scheurer E, Yen K, Fazekas F, Ropele S. Susceptibility induced gray-white matter MRI contrast in the human brain. Neuroimage. 2012;59(2):1413–1419. doi: 10.1016/j.neuroimage.2011.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou D, Cho J, Zhang J, Spincemaille P, Wang Y. Susceptibility underestimation in a high-susceptibility phantom: Dependence on imaging resolution, magnitude contrast, and other parameters. Magnetic resonance in medicine. 2017;78(3):1080–1086. doi: 10.1002/mrm.26475. [DOI] [PubMed] [Google Scholar]
- 60.Gardener AG, Jezzard P. Investigating white matter perfusion using optimal sampling strategy arterial spin labeling at 7 Tesla. Magnetic resonance in medicine. 2015;73(6):2243–2248. doi: 10.1002/mrm.25333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Table S1. CMRO2 (µmol/100g/min) and OEF (%) for the cortical gray matter for 11 subjects (Mean ± Standard Deviation)


