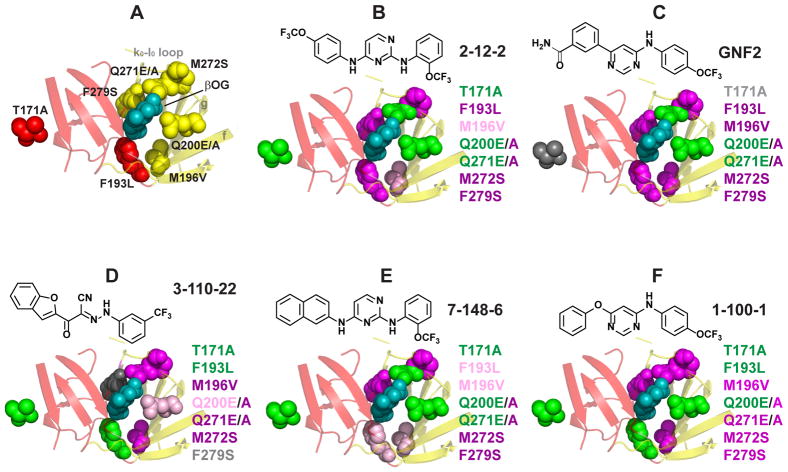
Figure 4. Loss-of-affinity footprints of representative DENV E inhibitors.
The ligand-binding pocket of the DENV2 E structure (1OKE) is shown with βOG (cyan) bound. (A) Substitutions were introduced at seven different residues lining the pocket as shown. Residues 200 and 271 were mutated from glutamine to glutamate (E) and alanine (A). (B–F) Loss-of-affinity for each mutation is mapped onto the 1OKE structure for cyanohydrazone 3-110-22, 2,4-diaminopyrimidines 2-12-2 and 7-148-6, and 4,6-disubstituted pyrimidines GNF2 and 1-100-1. Magenta, > 5-fold loss of affinity; light pink, 3- to 5-fold loss of affinity; green, < 2-fold loss of affinity; gray, data indeterminate due to poor signal-to-background. Color-coding at residue 271 reflect the effects of the Q271E mutation because the Q271A mutant exhibited significant loss of binding with all inhibitors.KD values are presented in Table S1.