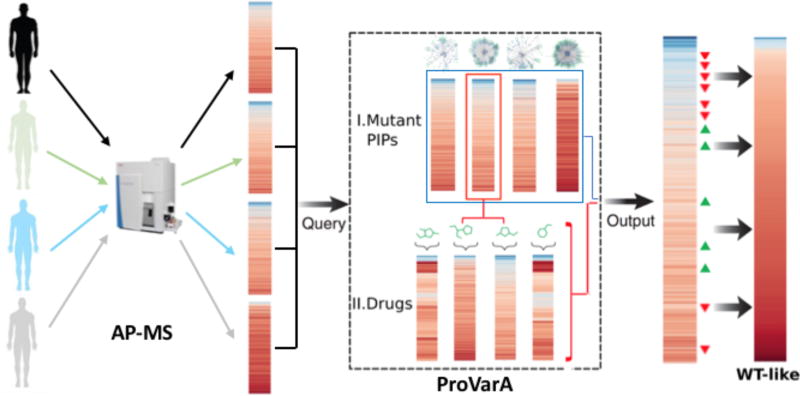
Figure 7. Workflow schematic for applying ProVarA to genetic diseases.
Schematic representation of how to apply ProVarA to human genetic diseases. In a first step a protein interaction profile (PIP) for the disease associated protein is generated from a patient. The resulting PIP is then queried against a database of established PIP for a panel of characterized disease-associated mutations in a 2-step process. The first step (I) aligns the patient or test PIP with the PIP of known mutations to identify target proteins that can be exploited for correction of the disease phenotype. In a second step (II) the response of the most similar reference PIP to a panel of small molecule therapeutics is mined to predict the most effective compounds for therapeutic intervention to restore a more WT-like PIP.