Fig. 3. Hepatic overexpression of DEPTOR ameliorates alcohol-mediated dysregulation of lipid metabolism in chronic-binge ethanol-fed mice.
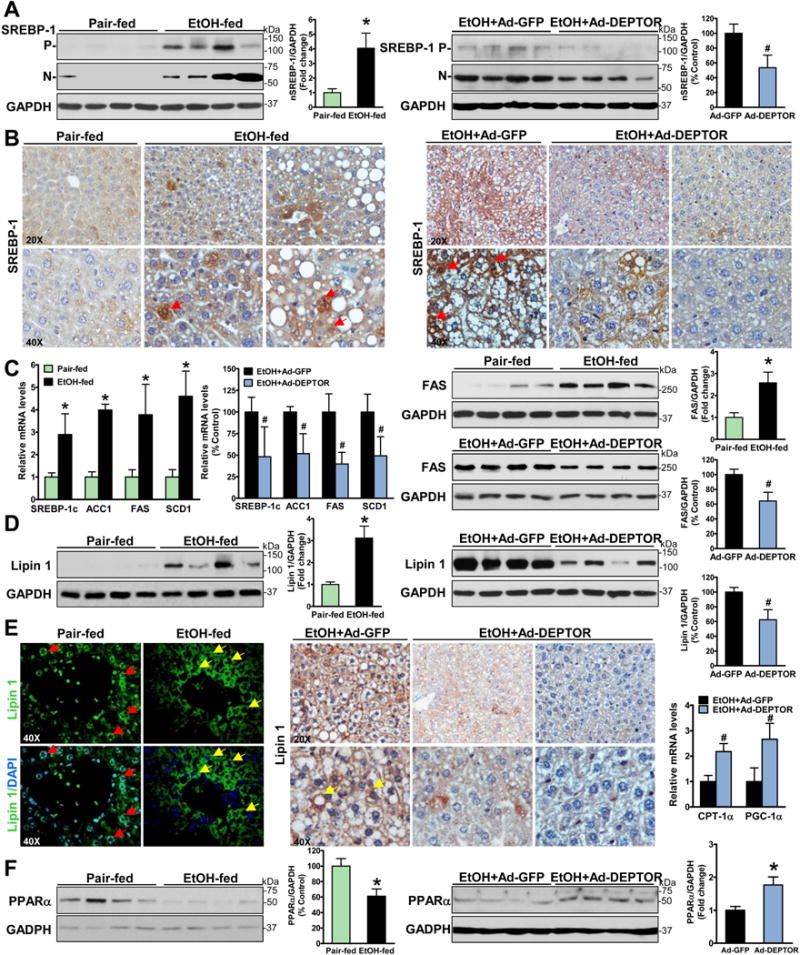
A. The active, nuclear form of SREBP-1 is increased in ethanol-fed mice and decreased by DEPTOR overexpression. P and N denote the precursor (~125 kDa) and cleaved nuclear (~68 kDa) forms of SREBP-1. B-C. The nuclear translocation of SREBP-1 as well as expression of SREBP-1c and its targets including ACC1, FAS, and SCD1 are reduced by DEPTOR overexpression. Notable, positive staining for SREBP-1 is primarily located in the nuclei of the hepatocytes of ethanol-fed mice (red arrows). D–E. Overexpression of DEPTOR represses the expression and cytoplasmic translocation of lipin-1 in chronic-binge ethanol-fed mice. Immunofluorescent staining showed that strong staining for lipin 1 (green) was predominantly located in the nuclear (blue) of hepatocytes (red arrows) of pair-fed mice, but it was elevated and mainly presented in the cytoplasm in hepatocytes (yellow arrows) surrounding central or portal veins of ethanol-fed mice. Notably, strong staining for lipin 1 is visualized mainly in the cytoplasm of hepatocytes (yellow arrows) of control mice on the ethanol diet, and this induction is inhibited by overexpression of DEPTOR. F. Expression of key genes involving fatty acid oxidation such as PPARα, CPT-1α and PGC-1α is analyzed. The data are presented as mean ± SEM, n = 6–8. *P < 0.05 vs. pair-fed mice. #P < 0.05 vs. ethanol-fed mice with Ad-GFP injection.
