Fig. 5. Hepatic S6K1 activity is required for the proteolytic activation of SREBP-1 and its lipogenic process in mice after chronic-binge ethanol feeding.
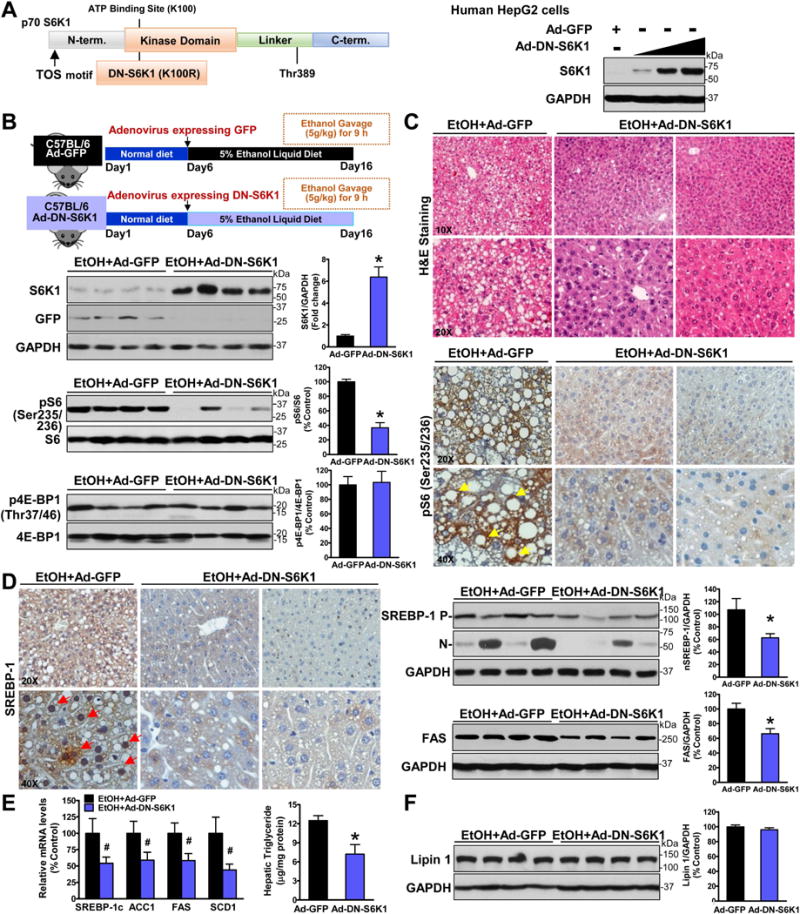
A. Schematic structure and phosphorylation sites of p70S6K1. The dominant negative form of S6K1 bears a mutation of lysine 100 to arginine (K100R) in the ATP binding site of the kinase domain. Immunoblots confirmed adenovirus-mediated overexpression of the dominant negative form of S6K1 (DN-S6K1) in HepG2 hepatocytes. B–C. Overexpression of DN-S6K1 inhibits S6K1 activity and attenuates hepatic steatosis in ethanol-fed mice. Notably, strong positive staining for phosphorylated S6 in the cytoplasm of hepatocytes (yellow arrows) in ethanol-fed mice is markedly decreased by overexpression of DN-S6K1. D–E. Overexpression of DN-S6K1 suppresses the accumulation of nuclear SREBP-1 and induction of lipogenic genes in response to ethanol feeding. Notably, positive staining for the nuclear translocation of SREBP-1 was presented in hepatocytes (red arrows) around central and portal veins in GFP-expressed mice. F. Alcohol-mediated induction of lipin 1 is unaffected by overexpression of DN-S6K1. The data are presented as mean ± SEM, n = 6–8. *P < 0.05 vs. ethanol-fed mice with Ad-GFP injection.
