Figure 2. Changes in transcription factor enrichment in response to T cell activation.
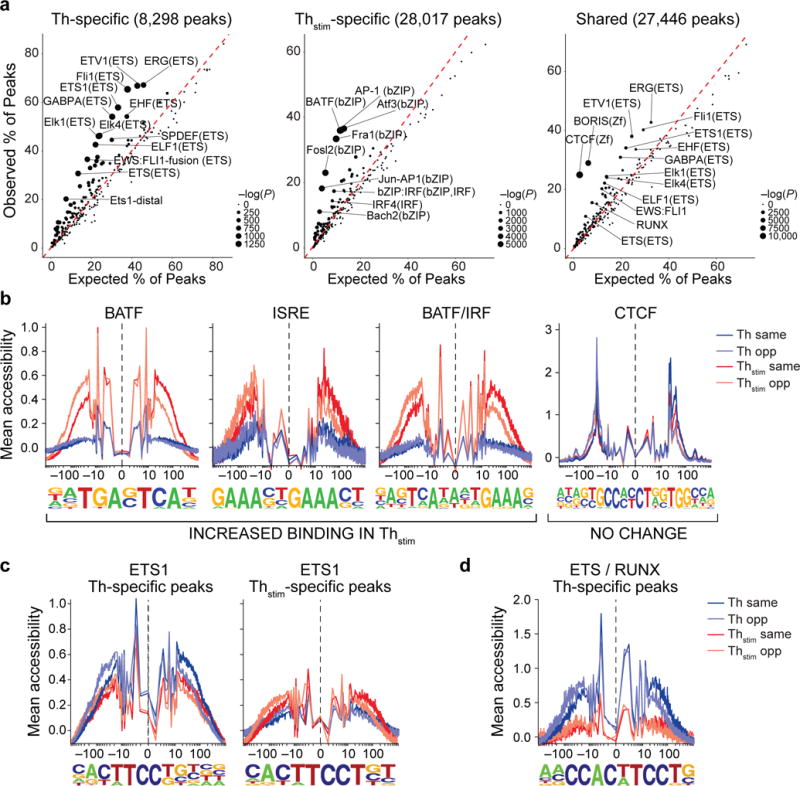
(a) Transcription factor motif enrichment. Expected (x-axis) vs. observed (y-axis) percentages of Th-specific (left), Thstim-specific (center), or shared (right) peaks overlapping each TF binding site annotation. (b-d) TF footprinting. For each TF motif (as defined in ENCODE63), nucleotide resolution average chromatin accessibility (y-axis) in Th (purple) or Thstim (red) cells along the TF binding site (x-axis; log(bp from center of each TF motif)). Aggregated locations are defined as (b) Thstim-specific peaks overlapping BATF, ISRE, and BATF/IRF motifs (three left panels) and shared peaks overlapping CTCF binding sites (right panel), (c) Th-specific (left) and Thstim-specific (right) peaks overlapping ETS1 binding sites, and (d) Th-specific peaks overlapping ETS1/RUNX combinatorial binding sites.
