Figure 5. Genetic determinants of co-accessible peaks.
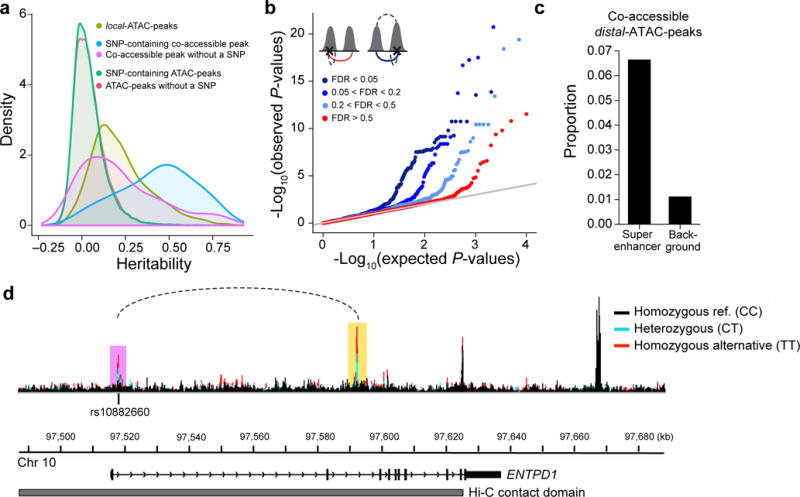
(a) Distribution of the heritability explained by SNPs +/− 500 kb of ATAC-peaks. Local-ATAC-peaks (olive). SNP-containing co-accessible peaks (blue). Co-accessible peaks without a SNP (purple). SNP-containing ATAC-peaks (green). ATAC-peaks without a SNP (red). (b) Q-Q plots of the linear regression P-values of distal-ATAC-peaks that are single peaks (red: co-accessibility FDR > 0.5), or co-accessible peaks called at various significance cutoffs (light blue: 0.2 < FDR < 0.5, medium blue: 0.05 < FDR < 0.2, dark blue: FDR < 0.05). The cartoons (upper left corner) depict the distal-ATAC-QTL association for single peaks (left cartoon; red line is the association plotted) and distal-ATAC-QTL association for co-accessible peaks (right cartoon; blue line is the association plotted; upper dashed line is the co-accessible peak at various significance cutoffs). (c) Proportion of co-accessible distal-ATAC-peaks overlapping super-enhancers or randomly shuffled background. (d) An example of a genetic variant (rs10882660) residing in the first intron in ENTPD1, associated locally (in purple) and distally (in yellow) to ATAC-peaks. The local and distal-ATAC-peaks are co-accessible (dotted line) and reside in a Hi-C contact domain (grey). ATAC-seq profiles were aggregated for individuals of different rs10882660 genotypes (black: homozygous major allele, light blue: heterozygous, red: homozygous minor allele).
