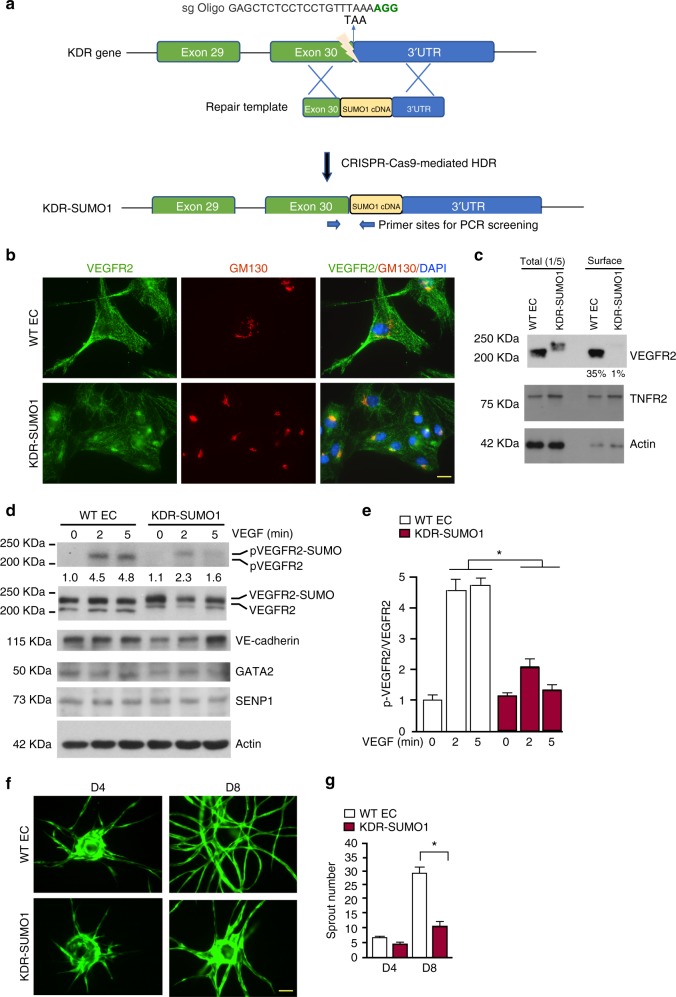
Fig. 6.
VEGFR2-SUMO1 fusion in human EC inhibits VEGF-mediated angiogenesis. a Schematic diagram of KDR-SUMO1 knockin in human EC by CRISPR/Cas9-mediated gene editing. A specific sgRNA targeting the vicinity of the stop codon of KDR gene encoding VEGFR2 and a repair template containing targeting arms with exon 30 as 5′ arm and the 3′UTR of the KDR gene as 3′ arm flanking the SUMO1 cDNA. Upon CRISPR/Cas9-mediated DNA double-strand breaks were repaired through homologous-directed repair, the SUMO1 cDNA was integrated into the KDR locus just before the termination signal. The PCR primers used for screening for KDR-SUMO1 fusion clones are indicated. b WT human EC and KDR-SUMO1 knockin EC were subjected to immunofluorescence staining for co-localisation of VEGFR2 with GM130. Similar pattern localisation was observed in cells derived from additional two clones. c WT human EC and KDR-SUMO1 knockin EC were cultured in normal media. Cell-surface VEGFR2 was labeled by cell-surface biotinylation, and analyzed by streptavidin bead pull-down followed by western blotting with anti-VEGFR2. Percentage of cell-surface VEGFR2 vs total VEGFR2 in WT EC and KDR-SUMO1 EC were quantified. n = 3. d, e WT human EC and KDR-SUMO1 knockin EC were treated with VEGF (10 ng/ml) for indicated times, and cells were subjected to western blot for VEGFR2 phosphorylation (c). Protein bands were quantified by densitometry and fold changes are presented by taking untreated WT group as 1.0 (d). n = 2. f, g 3D spheroid sprouting assay. Human ECs were infected with EGFP-expressing retroviruses. Cells were coated with microbeads, embedded in fibrin gels and grown in EGM2 medium for 8 days. A representative image of ten beads for each sample is shown (f). Quantification of sprout number is shown in panel (g). Three independent experiments were performed. Error bars, mean ± SEM; *P < 0.05, one-way ANOVA. Scale bar: 20 μm (b); 1 mm (f)