Figure 1.
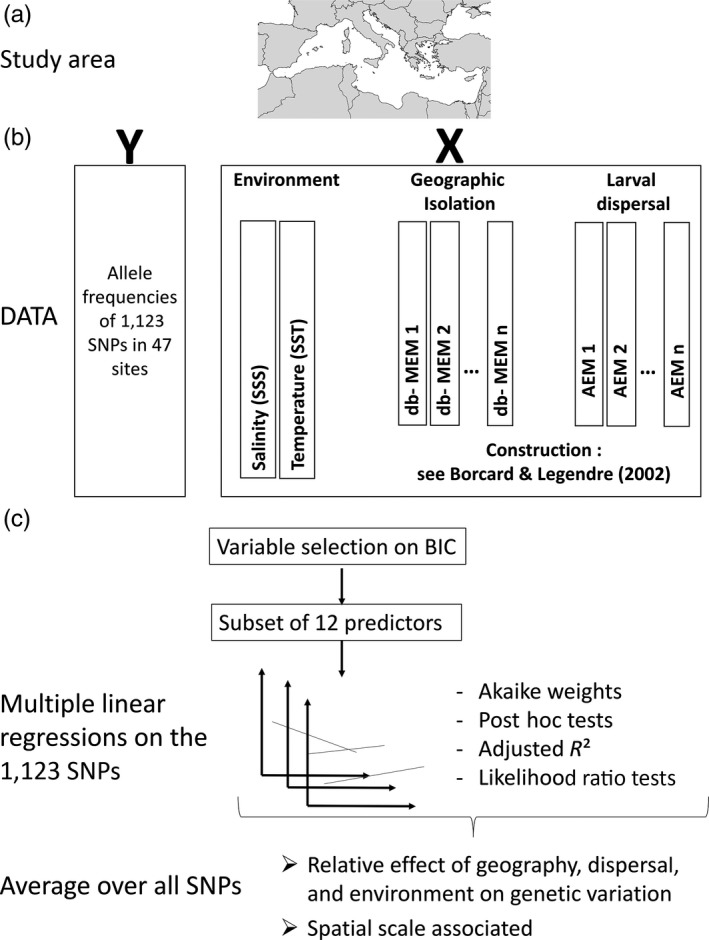
Analytical framework used to test the influence of environment, geographic isolation, and larval dispersal on the genetic variation of Mullus surmuletus and their associated spatial scales. (a) The study area, the Mediterranean Sea. (b) The response (SNP allele frequencies) and explanatory variables (environment, geographic isolation, and larval dispersal). Distance‐based Moran's eigenvector maps (db‐MEMs) were used to calculate node‐based predictors of geographic isolation. See Borcard and Legendre (2002) for more detailed explanation on the construction of db‐MEMs. Similarly, asymmetric eigenvector maps (AEMs) were used to calculate node‐based predictors of larval dispersal; construction of AEMs is detailed in Blanchet et al. (2008a). (c) Multiple linear regressions and the Akaike weight used to estimate the relative effect of each type of variable on variation in allele frequencies, and at which spatial scale they are mainly associated
