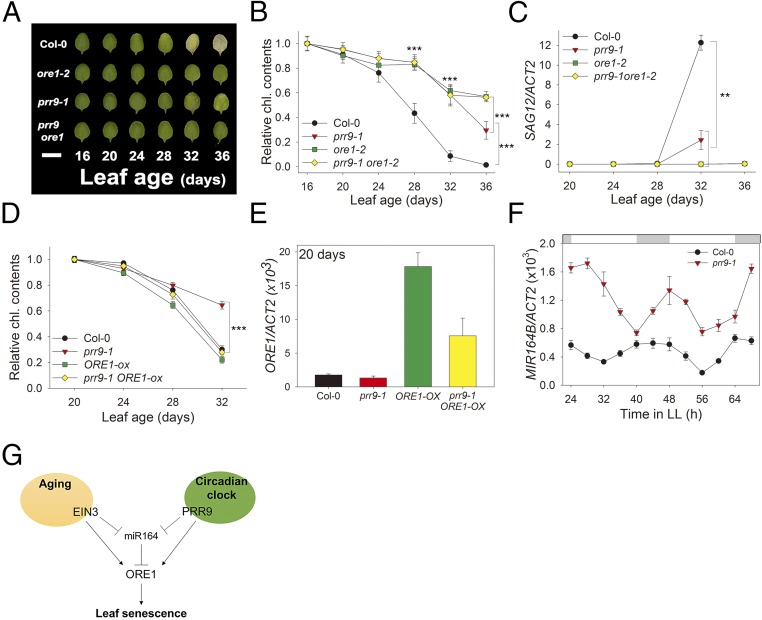
Fig. 5.
PRR9 positively regulates leaf senescence via ORE1. (A) Chlorophyll loss in Col-0, ore1-2, prr9-1, and the prr9-1ore1-2 double mutant. The photographs show representative third and fourth rosette leaves at the indicated days after leaf emergence. (Scale bar: 1 cm.) (B) The chlorophyll contents of the indicated genotypes were measured from the third and fourth leaves at indicated days (mean ± SE, n = 10). Asterisks indicate statistically significant difference from Col-0 (t test, ***P < 0.001). (C) The expression of SAG12 gene in plants of the indicated genotypes at the indicated days (mean ± SEM, n = 3). Asterisks indicate statistically significant difference from Col-0 (t test, **P < 0.01). (D) The chlorophyll content of the third and fourth leaves of the indicated genotypes was measured at indicated days (mean ± SEM, n = 8). Asterisks indicate statistically significant difference from Col-0 (t test, ***P < 0.001). (E) The expression of ORE1 gene in plants of the indicated genotypes at the indicated days (mean ± SEM, n = 3). (F) Level of MIR164B transcript in Col-0 and prr9-1 in 20-d-old leaves under LL (mean ± SEM, n = 3). ACT2, internal control. (G) A trifurcate feed-forward pathway model for regulating leaf senescence by aging and circadian clock. PRR9 positively regulates ORE1 with a circadian rhythm at both the transcriptional and posttranscriptional levels. Posttranscriptional repression by clock-controlled miR164 negatively regulates ORE1 mRNA level. Aging activates ORE1 expression by a similar trifurcate feed-forward pathway; direct binding of EIN3 to the ORE1 and miR164 promoters activates ORE1 expression directly and indirectly, via repression of miR164, a repressor of ORE1, to promote age-dependent senescence.