Fig. 4.
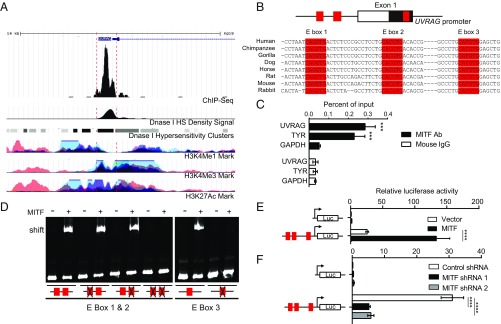
UVRAG is a transcriptional target of MITF. (A) University of California Santa Cruz genome browser view shows the 10-kb region around the UVRAG promoter. The ChIP-seq signals for MITF (30), and peaks of H3K4Me1, H3K4Me3, H3K27Ac, and DNase I hypersensitivity (HS) density in human melanocytes are extracted from the ENCODE dataset and annotated. (B) Structure of UVRAG promoter showing the location of exon 1 and the three E boxes of the MITF-binding sites (E-box 1, E-box 2, and E-box 3; in red) highly conserved through different species as indicated. (C) ChIP analysis shows MITF occupancy on the UVRAG promoter in human melanocytes. The TYR and GAPDH promoters were used as the positive and negative control, respectively. ***P < 0.001. (D) Electrophoretic mobility shift assay (EMSA) shows the direct association of purified recombinant MITF protein with three E-box sequences in the UVRAG promoter, but not with the E-box mutated sequences. EMSA probes containing either the WT or the mutant E-box sequence are indicated. (E) Dual luciferase reporter assay showing the activity of the UVRAG promoter in response to transfection of MITF in 293T cells. Red boxes indicate the location of three E boxes. Data are mean ± SD from three independent experiments. (F) Knockdown of MITF strongly suppresses UVRAG promoter activity in melanocytes. The UVRAG promoter–reporter was transfected into human melanocytes expressing MITF-specific shRNA and luciferase activity was measured 48 h posttransfection. Results are expressed as mean ± SD from three independent experiments. ****P < 0.0001.