Significance
The RNA interference (RNAi) discovered in nematodes has contributed to major advances in basic and applied sciences. RNAi-based methods are being developed for controlling pests and disease vectors. RNAi is highly efficient and systemic in coleopteran insects, but not in other insects. The lower efficiency of RNAi in economically important insects and concerns about resistance development are hindering the widespread use of this technology. To address these problems, a RNAi-sensitive Colorado potato beetle, Leptinotarsa decemlineata, and a cell line derived from this insect were used to identify a dsRNA-binding protein, StaufenC, as a major contributor to RNAi and its resistance. Interestingly, StaufenC homologs are present in only coleopteran insects and are essential for efficient RNAi response and its resistance in these insects.
Keywords: Tribolium, Leptinotarsa, RNAi efficiency, resistance, siRNA
Abstract
RNA interference (RNAi) is being used to develop methods to control pests and disease vectors. RNAi is robust and systemic in coleopteran insects but is quite variable in other insects. The determinants of efficient RNAi in coleopterans, as well as its potential mechanisms of resistance, are not known. RNAi screen identified a double-stranded RNA binding protein (StaufenC) as a major player in RNAi. StaufenC homologs have been identified in only coleopteran insects. Experiments in two coleopteran insects, Leptinotarsa decemlineata and Tribolium castaneum, showed the requirement of StaufenC for RNAi, especially for processing of double-stranded RNA (dsRNA) to small interfering RNA. RNAi-resistant cells were selected by exposing L. decemlineata, Lepd-SL1 cells to the inhibitor of apoptosis 1 dsRNA for multiple generations. The resistant cells showed lower levels of StaufenC expression compared with its expression in susceptible cells. These studies showed that coleopteran-specific StaufenC is required for RNAi and is a potential target for RNAi resistance. The data included in this article will help improve RNAi in noncoleopteran insects and manage RNAi resistance in coleopteran insects.
Exposure of cells to double-stranded RNA (dsRNA) causes target-specific gene silencing known as RNA interference (RNAi). RNAi was discovered in the nematode, Caenorhabditis elegans, and was subsequently observed in most eukaryotes including humans, plants, and insects (1). dsRNAs synthesized in vitro or in microorganisms (including bacteria, yeast, and algae) or plants have been used to achieve gene silencing in insects (2–4). RNAi-aided gene silencing methods helped to determine functions of genes identified by the genome, transcriptome, and proteome sequencing and advance our understanding of the molecular basis of many developmental and physiological processes including insect development, reproduction, behavior, and communication (5, 6). RNAi technology is being employed to develop methods to control crop pests as well as vectors that transmit deadly diseases affecting humans, animals, and plants (2–4, 7, 8). RNAi is also used to identify target sites for insecticide development, as well as to manage insecticide resistance in pests and disease vectors (7–9). The US Environmental Protection Agency recently approved transgenic corn plants expressing dsRNA for the control of Western corn rootworm in the United States (https://www.regulations.gov/docket?D=EPA-HQ-OPP-2014-0293).
RNAi efficiency is variable among insects tested so far. RNAi works very well and is systemic in beetles (Coleoptera), including the red flour beetle (Tribolium castaneum), Colorado potato beetle (Leptinotarsa decemlineata), and Western corn rootworm (Diabrotica virgifera virgifera) (7–10). In contrast, RNAi works poorly in moths and butterflies belonging to the order Lepidoptera (11, 12). Differences in digestion of dsRNA by endogenous dsRNases (dsRNA endonucleases), in transport of dsRNA into cells and trafficking within cells, in processing of dsRNA to siRNA, and in expression of RNAi genes and composition of proteins coded by these genes have been identified as the main contributors to differential RNAi efficiency among insects (13–16). Recent studies identified dsRNA digestion by dsRNases and entrapment of dsRNA in endosomes as the major contributors to RNAi recalcitrance in lepidopteran insects and cell lines (17). Whether or not any coleopteran insect-specific proteins contribute to robust RNAi in these insects is not known.
Cells defend themselves from dsRNAs from many sources (e.g., viruses). The enzyme Dicer-2 cleaves dsRNAs to small interfering RNAs (siRNAs), which are then incorporated into an RNA-induced silencing complex, bind to complementary mRNAs, and interfere with their translation (18, 19). Key proteins including Dicer-2, R2D2, Loquacious (Loqs), and Argonautes involved in RNAi pathway have been identified (20–22). However, the difference in composition and function of silencing complexes between insects in which RNAi works efficiently (e.g., coleopterans) and those in which RNAi does not work efficiently (e.g., lepidopterans) remains unknown.
Staufen was discovered in Drosophila melanogaster in a screen for maternal effect mutations (23). Molecular genetic studies have shown that D. melanogaster Staufen is required to localize mRNAs in oocytes and neuroblasts (24, 25). Human and mouse Staufen homologs were identified and the human staufen gene codes for two proteins (Stau1 and Stau2) with differences in the N-terminal end (26). Staufen1 is the mammalian homolog of D. melanogaster Staufen and is thought to function in mRNA transport, translational control, and mRNA decay (27–29). The staufen protein contains multiple double-stranded RNA binding domains, and some of them are shown to bind dsRNA (26, 30).
Insects have developed resistance to almost all synthetic chemicals used for their control, resulting in a constant battle between humans and insects. It is possible that insects will develop resistance to RNAi-based technologies as well. L. decemlineata has developed resistance to almost all synthetic chemicals used for its control within a short period of 2–3 y after their introduction (31). Therefore, L. decemlineata could be a good model insect to study potential RNAi resistance. Resistance in dsRNA-treated insects might be developed by the selection of individuals with modification in genes coding for proteins functioning in the RNAi pathway. Alternatively, insects might develop resistance to dsRNAs by selection of individuals with mutations in the dsRNA target sites. Information on potential mechanisms of RNAi resistance is needed to make progress in the widespread use of RNAi for controlling insect pests and disease vectors. We used L. decemlineata and a cell line derived from this insect to identify proteins required for RNAi in coleopteran insects, as well as to study potential mechanisms of RNAi resistance.
Results
StauC Is a Major Contributor to RNAi.
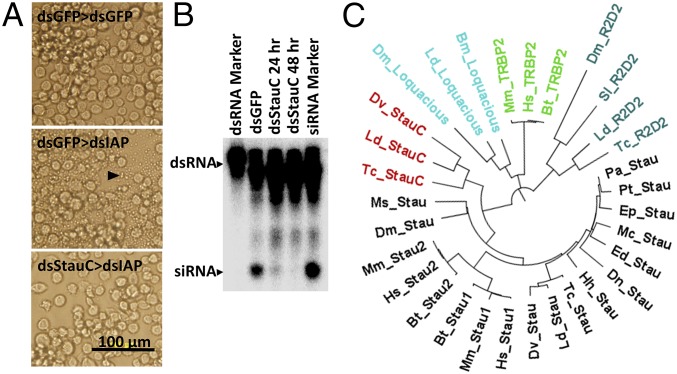
In a recent study, we screened 50 genes in Lepd-SL1 cells and identified five genes (Argonaute-1, Argonaute-2a, Argonaute-2b, Aubergine, and vha16) that are essential for RNAi (32). In these experiments, when Lepd-SL1 cells were exposed to StaufenC (named as StauC because of its presence only in coleopteran insects) dsRNA for 24 h, followed by a second dsRNA targeting the gene coding for inhibitor of apoptosis 1 (IAP), apoptosis was detected in some of the cells. Therefore, StauC was not selected as a gene essential for RNAi response in these experiments (32). However, in subsequent experiments, when the dsStauC pretreatment was increased to 48 h, no apoptosis was detected in the cells exposed to dsIAP, suggesting StauC is required for RNAi in Lepd-SL1 cells (Fig. 1A). We hypothesized that StauC RNA and/or protein might be more stable than the other gene products, and therefore, a longer period of dsRNA treatment is required to deplete StauC from these cells. To determine whether StauC is required for processing of dsRNA to siRNA in Lepd-SL1 cells, 32P-labeled dsGFP was used to track dsRNA processing in cells exposed to dsStauC or dsGFP (dsRNA targeting the gene coding for the green fluorescent protein, dsGFP, used as a control). The control Lepd-SL1 cells exposed to 32P-labeled dsGFP processed dsRNA to siRNA (Fig. 1B). However, the RNA isolated from the cells exposed to dsStauC for 24 or 48 h showed a faint siRNA band and no band, respectively (Fig. 1B). The siRNA detected in cells exposed to dsStauC for 24 h is at a higher intensity than that detected in cells exposed to dsStauC for 48 h, confirming our hypothesis that complete knockdown of StauC requires up to 48 h of exposure to dsRNA (Fig. 1B). These data show that knockdown of StauC negatively affect dsRNA-to-siRNA processing in these cells.
Fig. 1.
Coleopteran-specific StauC is a major contributor to RNAi pathway. (A) Phenotypes of cells exposed to dsGFP or dsStauC followed by exposure to second dsRNA (dsGFP or dsIAP). The Lepd-SL1 cells were exposed to the first dsRNA (dsGFP or dsStauC) for 48 h, followed by the treatment with the second dsRNA (dsGFP or dsIAP). Photographs were taken at 24 h after treatment with the second dsRNA. The arrow points to the cells undergoing apoptosis. (B) Comparison of processing of dsRNA to siRNA in Lepd-SL1 cells exposed to dsGFP (control) or dsStauC. The Lepd-SL1 cells seeded in six-well plates were incubated with dsGFP or dsStauC for 24 or 48 h. Then, the cells were exposed to 1.6 million cpm 32P-labeled dsGFP. At 48 h after the addition of the second dsRNA, the cells were harvested, and total RNA was isolated and the RNA containing 2,000 cpm was resolved on 16% acrylamide-urea gel. The gel was dried and analyzed using a phosphorImager. The first lane shows GFP dsRNA used as a marker, and the last lane shows dsRNA processed to siRNA in Lepd-SL1 cells used as a marker for siRNA. The arrows point to dsRNA and siRNA bands. (C) Phylogenetic tree of dsRNA-binding proteins. Major clusters of proteins include Staufen (mammalian Stau1 and Stau2, and insect Staufens from 10 insect orders, and coleopteran StaufenC), insect dsRNA binding proteins (R2D2 and Loquacious), and mammalian RNA binding protein (TRBP2). The sequences of Staufen (Megaloprepus caerulatus, Periplaneta americana, Empusa pennata, L. decemlineata, D. melanogaster, Manduca sexta, Dufourea novaeangliae, Ephemera danica, Prosarthia teretrirostris, D. virgifera virgifera, and Halyomorpha halys), StauC (L. decemlineata, T. castaneum, and D. virgifera virgifera), Stau1 (Bos taurus, Mus musculus, and Homo sapiens), Stau2 (B. taurus, M. musculus, H. sapiens), Loquacious (D. melanogaster, Bombyx mori, L. decemlineata), R2D2 (L. decemlineata, T. castaneum, D. melanogaster, Spodoptera litura), and TRBP2 (H. sapiens, B. taurus, and M. musculus) are included in the analysis. The Muscle program in MEGA 7.0 was used to align the protein sequences, and the maximum likelihood analysis was performed with bootstrapping (1,000 replicates).
StauC Gene Sequences Are Present in only Coleopteran Insects; StauC Is Required for RNAi in Beetles.
Blast searches were conducted to identify StauC homolog sequences deposited in the GenBank and i5K databases. Staufen homolog sequences were identified in most of the insect genome and transcriptome databases searched. However, we were not able to find any StauC homolog sequences in insects outside the order Coleoptera. Twenty-three of 32 coleopteran insect genomes/transcriptomes searched showed two Staufens: Staufen (Stau, present in all insects and contains conserved Stau sequence and four RNA binding domains; SI Appendix, Fig. S1) and StaufenC (StauC, present only in insects from Coleoptera and contains conserved Stau sequence and three RNA binding domains; SI Appendix, Fig. S1). Eight of 32 coleopteran genome and transcriptome databases searched showed only Stau sequences (SI Appendix, Fig. S2). In addition, we could find StauC, but not Stau, sequence in the cotton boll weevil, Anthonomous grandis, transcriptome (SI Appendix, Fig. S2). StauC sequences are not found in the genomes of other organisms including mammals. Phylogenetic analysis placed Stau sequences from mammals (Stau1 and Stau2) and insects in a group that did not include StauC and other dsRNA-binding proteins (Fig. 1C). Interestingly, unlike StauC, Stau is not required for RNAi response in Lepd-SL1 cells (SI Appendix, Fig. S3).
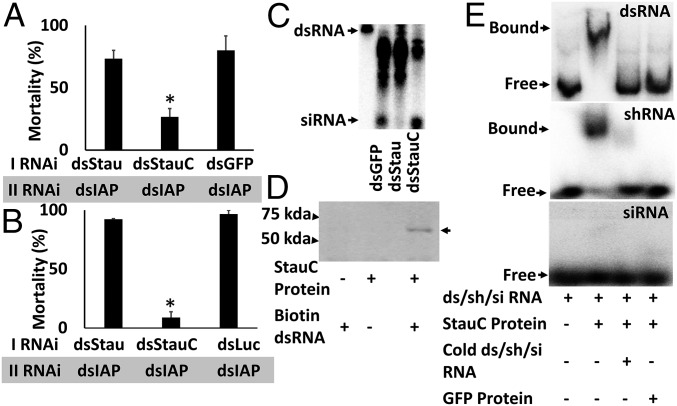
To determine whether StauC has a conserved role in efficient RNAi in vivo in L. decemlineata and other coleopteran insects, dsStauC, dsStau, dsGFP, or dsLuc (dsRNA targeting the luciferase gene used as a control) was injected into L. decemlineata or T. castaneum larvae. After 2–3 d, these larvae were fed or injected with a second dsRNA targeting the gene coding for IAP. As shown in Fig. 2 A and B, treatment with StauC dsRNA followed by exposure to dsIAP resulted in significantly less mortality compared with that in control larvae treated with dsGFP/dsLuc followed by dsIAP. Interestingly, the mortality observed in dsStau-injected larvae is similar to the mortality in the control larvae. These data show that StauC (but not Stau) is required for RNAi in L. decemlineata and T. castaneum. Therefore, it appears that although coleopteran insects code for two Staufen proteins, only one of them, StauC, is required for RNAi response.
Fig. 2.
StauC is essential for RNAi in L. decemlineata and T. castaneum. (A) In vivo RNAi in L. decemlineata. The third instar L. decemlineata larvae were injected with 1,000 ng Stau, StauC, or GFP (control) dsRNA. Three days later, the larvae were fed on leaf discs treated with 25 ng dsIAP. The mortality was recorded until the control larvae reached the pupal stage. Mean + SE (n = 15) are shown. *Significantly different from control, at P ≤ 0.05. (B) In vivo RNAi in T. castaneum. The last instar T. castaneum larvae were injected with 200 ng Stau, StauC, or Luc (control) dsRNA. At 48 h after first dsRNA injection, 200 ng dsIAP was injected and the mortality was recorded until the control larvae reached the pupal stage. Mean + SE (n = 15) are shown. *Significantly different from control at P ≤ 0.05. (C) StauC is required for dsRNA processing to siRNA in vivo. One microgram dsGFP, dsStau, or dsStauC was injected into each second instar L. decemlineata larva. Three days after injection, each larva was fed on 2 million cpm dsGFP. At 72 h after feeding dsRNA, total RNA was isolated and the RNA containing 2,000 cpm was resolved on 16% acrylamide-urea gel. The gel was dried and analyzed using a phosphorImager. The first lane shows GFP dsRNA used as a marker for dsRNA. The arrows point to dsRNA and siRNA bands. (D) StauC binds to dsRNA. Purified StauC protein was mixed with biotinylated dsGFP, and the mixture was pulled down using streptavidin beads. The same amount of BSA and unlabeled dsGFP were used as negative controls. The eluted sample was resolved on SDS/PAGE (10%) gel and transferred to the membrane, and StauC antibody was used to detect StauC on Western blots. The position of 75- and 50-kDa proteins run on the same gel are shown on the left. (E) StauC expressed in E. coli binds to 32P-labeled dsGFP. The labeled dsRNA and StauC protein or StauC protein plus 100× unlabeled dsRNA/shRNA/siRNA or GFP protein were added to the gel shift reaction and incubated for 20 min. Then the complexes were resolved on nondenaturing PAGE (4%). The gel was dried and analyzed using a phosphorImager.
To confirm the involvement of StauC in dsRNA processing in vivo, L. decemlineata larvae were injected with dsGFP, dsStauC, or dsStau dsRNA followed by feeding on 32P-labeled dsGFP. As shown in Fig. 2C, dsRNA was processed to siRNA in control larvae treated with dsGFP or the larvae treated with dsStau, but not in larvae treated with dsStauC. These data show that StauC is required for RNAi in vivo and confirm results from the experiments using the Lepd-SL1 cell line and suggest that StauC present only in coleopteran insects is a major contributor to the robust RNAi response observed in this group of insects.
StauC Binds to dsRNA and shRNA.
Analysis of StauC sequences from 24 coleopteran insects identified four conserved dsRNA binding domains including Staufen domain (SI Appendix, Fig. S1). To determine whether StauC from L. decemlineata can bind dsRNA, we performed pull-down and gel mobility shift assays. StauC bound to biotinylated GFP dsRNA, as shown by precipitation of StauC/dsRNA complex by streptavidin, followed by identification of StauC protein by Western blot hybridization using StauC polyclonal antibodies (Fig. 2D). StauC expressed in Escherichia coli bound to 32P-labeled dsRNA (dsGFP) and short hairpin RNA targeting the luciferase gene (shLuc), and the binding was competitively inhibited by excess cold dsGFP or shLuc, demonstrating the specific binding of StauC to dsRNA or shRNA (Fig. 2E). Interestingly, StauC protein did not bind to 32P-labeled siRNA (Fig. 2E). These data suggest that StauC binds to long dsRNA and shRNA, but not to siRNA.
StauC Is Involved in Dicer-2 Processing of dsRNA to siRNA.
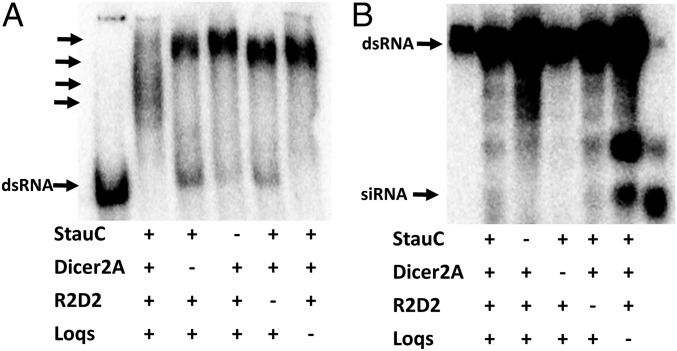
Experiments in L. decemlineata cells and in vivo suggest that StauC plays a key role in the processing of dsRNA to siRNA. To determine whether StauC is involved in the Dicer-2 processing of dsRNA to siRNA, we prepared embryonic extracts using eggs laid by females injected with StauC, Dicer-2a, R2D2, or Loqs dsRNA and incubated them with 32P-labeled dsRNA, and the products were resolved by nondenaturing or denaturing PAGE. The embryonic extract from eggs laid by control females formed multiple dsRNA–protein complexes (Fig. 3A). However, these complexes were not formed when embryonic extracts prepared from eggs laid by females injected with Dicer-2a, StauC, R2D2, or Loqs dsRNA were used. A single band running close to the origin was detected (Fig. 3A). The dsRNA–embryonic protein complexes resolved on denaturing gels showed a typical siRNA band when embryonic extracts from eggs laid by control females and females treated with R2D2 or Loqs dsRNA were used (Fig. 3B). The siRNA band was not detected when embryonic extracts from eggs laid by females injected with StauC or Dicer-2a dsRNA were used (Fig. 3B). These data suggest that StauC may be involved in the formation of dsRNA–Dicer-2a RNAi initiator complexes for processing of dsRNA to siRNA.
Fig. 3.
StauC is required for processing of dsRNA to siRNA. (A) Newly emerged female L. decemlineata were injected with 1 µg GFP, StauC, Dicer-2a, R2D2, or Loqs dsRNA. Ten days after injection, the females were mated with male beetles. Freshly laid eggs were collected, embryonic extracts were prepared, and the protein concentration was determined. A mixture of lysate, 32P-labeled dsRNA, and the reaction mixture containing ATP were incubated at 25 °C for 3 h. Then the reaction was deproteinized by adding proteinase K and the RNA was precipitated by adding 3 M sodium acetate and 3 volumes of absolute ethanol. The RNA was resolved on 16% acrylamide-urea gel. The gel was dried and analyzed using a phosphorImager. The first lane shows the labeled GFP as the marker for dsRNA. The arrows point to dsRNA complexes. (B) Embryonic lysate prepared as described in the legend of A, 32P-labeled dsRNA and the reaction mixture containing ATP were incubated at 25 °C for 1 h. The RNA was resolved on nondenaturing 4% acrylamide gel. The gel was dried and analyzed using a phosphorImager. The first lane shows GFP dsRNA used as a marker, and the last lane shows dsRNA digested to siRNA with RNase III and used as a marker for siRNA. The arrows point to dsRNA and siRNA bands.
Selection of RNAi-Resistant Lepd-SL1 Cells and Identification of Resistance Mechanisms.
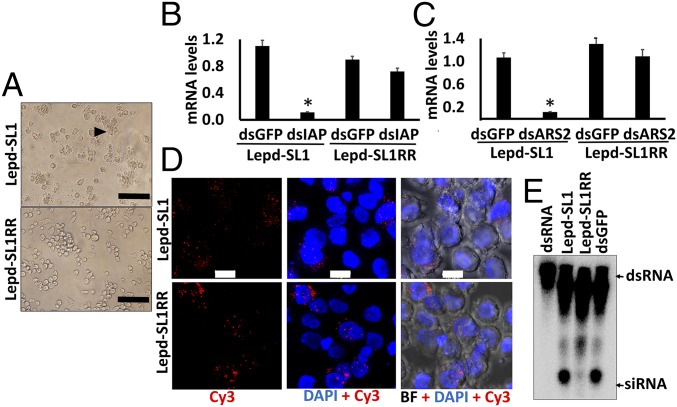
Inhibitor of apoptosis proteins (IAP), first discovered in baculoviruses and later in multicellular organisms, including insects, protects the cells from programmed cell death (33). We took advantage of the property of this protein to design a strategy for selecting coleopteran cells that are resistant to RNAi. Exposure of Lepd-SL1 cells to increasing concentration of dsIAP, followed by culture of surviving cells for multiple rounds, resulted in the selection of Lepd-SL1 RNAi-resistant cells (Lepd-SL1RR). Exposure of Lepd-SL1 cells to 20 ng dsIAP per well in a 96-well plate for 24 h induced apoptosis in many cells (Fig. 4A). In contrast, Lepd-SL1RR cells exposed to the same concentration of dsIAP for 24 h showed apoptosis in only a few cells (Fig. 4A). Also, treating Lepd-SL1 and Lepd-SL1RR cells with different concentrations of dsRNA (2.5–20 ng) showed similar differences in RNAi response (SI Appendix, Fig. S4). To determine whether the lack of apoptosis in Lepd-SL1RR cells is a result of a reduction in knockdown of IAP, we determined relative IAP mRNA levels in Lepd-SL1 and Lepd-SL1RR cells exposed to dsIAP and a control dsGFP. As shown in Fig. 4B, a significant decrease in IAP mRNA levels was detected in Lepd-SL1 cells, but not in Lepd-SL1RR cells exposed to dsIAP. These data suggest that knockdown of the IAP gene is significantly lower in Lepd-SL1RR cells exposed to dsIAP compared with that in Lepd-SL1 cells. To determine whether the lack of knockdown of the IAP gene in dsIAP exposed Lepd-SL1RR cells is specific to dsIAP or whether these cells are resistant to dsRNA-mediated gene knockdown in general, we compared the knockdown efficiency of two additional genes in these two cell lines. The mRNA levels of Arsenate resistance protein 2 (ARS2) and Protein arginine N-methyltransferase 5 (PRMT5) were determined in Lepd-SL1 and Lepd-SL1RR cells exposed to dsARS2, dsPRMT5, or dsGFP. A significant knockdown in both ARS2 and PRMT5 genes was observed in Lepd-SL1 cells exposed to corresponding dsRNAs (Fig. 4C and SI Appendix, Fig. S5). In contrast, no significant reduction of ARS2 or PRMT5 mRNA levels was detected in Lepd-SL1RR cells exposed to ARS2 or PRMT5 dsRNAs (Fig. 4C and SI Appendix, Fig. S5). These data suggest that the lack of RNAi response detected in Lepd-SL1RR cells is not specific to the IAP gene or mutations in the IAP gene target region but, rather, is likely a result of changes in RNAi pathway genes in these cells. To investigate which RNAi pathway component is changed in the resistant cells, we first checked whether dsRNA could be successfully taken up by the Lepd-SL1RR cells. Both Lepd-SL1 and Lepd-SL1RR cells were exposed to Cy3-labeled dsGFP. The cells were fixed at 2 h after treatment and observed under a confocal microscope. Both Lepd-SL1 and Lepd-SL1RR cells showed internalized labeled dsRNA, suggesting that transport of dsRNA into resistant cells is unlikely to be the main contributor to RNAi resistance in these cells (Fig. 4D and SI Appendix, Fig. S6A). No fluorescence was detected inside the cells exposed to Cy3 dye alone (SI Appendix, Fig. S6B). The Cy3-labeled dsIAP is able to induce apoptosis in Lepd-SL1 cells, demonstrating that labeling did not affect its function (SI Appendix, Fig. S6C).
Fig. 4.
Development of the RNAi-resistant Lepd-SL1 cell line and identification of resistance mechanisms. (A) Apoptosis phenotype observed after exposing Lepd-SL1 and Lepd-SL1RR cells to dsIAP. The cells were exposed to 20 ng dsIAP in 100 µL medium and photographed at 24 h after treatment. The arrow points to the cells showing apoptosis phenotype. (Scale bar, 100 μm.) (B) Relative IAP mRNA levels in Lepd-SL1 and Lepd-SL1RR cells exposed to dsIAP. The cells were exposed to dsIAP or dsGFP (a control), and total RNA was isolated and used to quantify relative IAP mRNA levels by qRT-PCR. Ribosomal protein 4 (RP4) was used as an internal control. The bars show mean ± SD (n = 3). *Significantly different from control at P ≤ 0.05. (C) Relative Ars2 mRNA levels in Lepd-SL1 and Lepd-SL1RR cells exposed to dsArs2 quantified as described in B legend. (D) Subcellular localization of Cy3-labeled dsGFP in the Lepd-SL1 and Lepd-SL1RR cells. The cells were exposed to 25 ng Cy3-labeled dsGFP in 8-well chamber slides for 2 h. Then the cells were fixed, mounted in DAPI containing medium and photographed using a confocal microscope. BF, bright field; Cy3, Cy3-labeled dsGFP. (Scale bar, 10 μm.) (E) Comparison of processing of dsRNA to siRNA in Lepd-SL1 and Lepd-SL1RR cells. The cells in six-well plates were exposed to 1.6 million cpm 32P-labeled dsGFP. At 24 h after the addition of dsRNA, the cells were harvested and RNA was isolated. RNA containing 2,000 cpm was resolved on 16% acrylamide-urea gel. The first and last lanes show GFP dsRNA and GFP dsRNA digested to siRNA with RNase III, respectively, used as markers. The arrows point to dsRNA and siRNA bands.
To confirm that both resistant and susceptible cells take up dsRNA and to track processing of dsRNA to siRNA, we exposed Lepd-SL1 and Lepd-SL1RR cells to 32P-labeled dsGFP, and the total RNA was isolated from the cells collected at 24 h after exposure to the labeled dsRNA. dsRNA bands were detected in the RNA isolated from both Lepd-SL1 and Lepd-SL1RR cells (Fig. 4E), demonstrating that both susceptible and resistant cells take up dsRNA. As reported previously (16), dsRNA was processed into siRNA in Lepd-SL1 cells (Fig. 3E). However, very little processed siRNA was detected in Lepd-SL1RR cells. Conversion of dsRNA to siRNA in Lepd-SL1RR cells was decreased by more than 80% compared with Lepd-SL1 cells. (Fig. 4E).
Lower Levels of StauC Expression in RNAi-Resistant Cells.
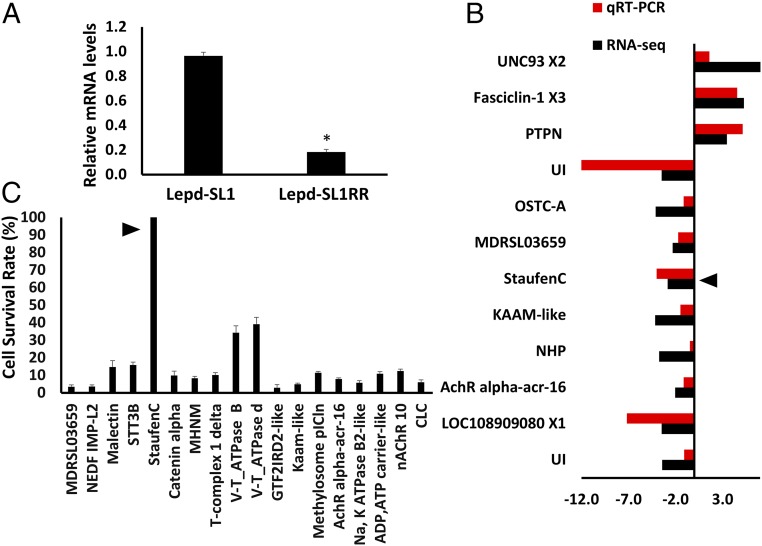
To identify genes whose expression changes affect the RNAi response in Lepd-SL1RR cells, the relative mRNA levels of 50 genes thought to be involved in RNAi (32) were determined by qRT-PCR in both Lepd-SL1 and Lepd-SL1RR cells. Among the 50 genes tested, only one gene, StauC, showed a more than twofold decrease in mRNA levels in Lepd-SL1RR cells compared with its levels in Lepd-SL1 cells (Fig. 5A and SI Appendix, Fig. S7). These data suggest that reduction in expression of StauC may be a significant contributor to RNAi resistance in Lepd-SL1RR cells. To determine whether there are any changes in expression of other genes besides StauC, we compared transcriptomes of Lepd-SL1 and Lepd-SL1RR cells (RNA sequences were deposited into the NCBI sequence read archive database under accession number SRP150964). Differential gene expression analysis identified 278 genes (104 down-regulated and 174 up-regulated) that are differentially expressed by twofold or more with a P value of ≤0.05 between resistant and susceptible cells (SI Appendix, Fig. S8 and Datasets S1 and S2). Interestingly, StauC is in the group of genes that are expressed more than twofold less in Lepd-SL1RR cells. The differential expression of a dozen genes randomly selected (nine down-regulated and three up-regulated) was confirmed by qRT-PCR (Fig. 5B). To determine whether the 18 genes that showed differential expression between resistant and susceptible cells (selected on the basis of their predicted function in dsRNA transport and processing) are required for RNAi response, the Lepd-SL1 cells were first exposed to dsRNA targeting each one of these genes, followed by dsIAP. Of the 18 genes tested, only knockdown of StauC resulted in 100% survival of cells exposed to dsIAP, suggesting that StauC is required for RNAi response (Fig. 5C). Knockdown of two other genes tested, V-ATPase B and V-ATPase d, resulted in 30% survival of cells, suggesting that these proteins may also contribute to RNAi (Fig. 5C). These data showed that a reduction in StauC mRNA levels is a key factor in the development of resistance to dsRNA in Lepd-SL1RR cells. A few other genes known to function in dsRNA transport and processing showed a change in their expression levels between Lepd-SL1 and Lepd-SL1RR cells. However, the mRNA levels of StauC are reduced to the greatest extent (SI Appendix, Fig. S8). In mammalian cells, mRNA Editase-related genes (ADAR1/Editase/Deaminase) are shown to mediate resistance to RNAi (34). None of these genes showed differential expression between Lepd-SL1 and Lepd-SL1RR cells (SI Appendix, Fig. S9), suggesting that the mechanisms of resistance could be different between mammalian and insect cells.
Fig. 5.
StauC is the major contributor to RNAi resistance. (A) Relative StauC mRNA levels in Lepd-SL1 and Lepd-SL1RR cells. Among the 50 RNAi genes tested by qRT-PCR, StauC is the only gene that showed more than twofold difference in expression between Lepd-SL1 and Lepd-SL1RR cells (details are in SI Appendix, Fig. S7). *Significantly different at P ≤ 0.05. (B) Comparison of differential expression of select genes between Lepd-SL1 and Lepd-SL1RR cells measured by qRT-PCR and RNA-seq. The arrow points to the StauC gene, Ld_c7641. Complete names of genes are listed in SI Appendix, Table S2. (C) Testing of 18 differentially expressed genes by RNAi assay, as described in the Fig. 1A legend. Complete names of genes are listed in SI Appendix, Table S3.
Discussion
The most important finding of this article is the identification of coleopteran-specific StauC as a critical player in robust and systemic RNAi response in these insects. After its discovery in D. melanogaster, Staufen homologs have been identified in both vertebrate and invertebrate animals. dsRNA binding property of Staufen suggests it might function in dsRNA-triggered RNAi response. To date, only the Staufen homolog identified from C. elegans was reported to be involved in RNAi response. Mutants of nematode Staufen showed enhanced RNAi response (35). This is exactly opposite to what we observed in the current study. Knockdown of StauC in L. decemlineata and T. castaneum severely impaired RNAi response. Interestingly, unlike in D. melanogaster and many other insects, beetles including L. decemlineata and T. castaneum genomes code for two Staufens, Stau and StauC. Among the coleopteran insects, we found StauC sequences in 24 of 32 genomes/transcriptomes searched. StauC sequence is found in only one of the four suborders (Polyphaga) of the order Coleoptera. Also, StauC sequence is present in nine of the 11 (10 from Polyphaga and one from Adephaga) beetle genome sequences available from the National Center for Biotechnology Information (NCBI) database. We did not find StauC sequence in Agrilus planipennis (Polyphaga) and Pogonus chalceus (Adephaga) genome sequences available from the NCBI. As pointed out in a recent review (36), complete genome sequences are lacking for most taxonomic groups in Coleoptera. Given the uncertainty in relationships among the higher taxa of this order, it is difficult to infer evolutionary transitions between Stau and StauC. Further studies are required to understand the origin and distribution of StauC in coleopteran insects.
The Staufen functions in RNA trafficking, decay, and translation repression in mammals (27–29). It is possible that StauC in coleopteran insects may be involved in intracellular trafficking of dsRNA to the sites for Dicer action. Our data showed that StauC is involved in formation of RNAi initiator complexes by bringing dsRNA and Dicer-2 together for processing of dsRNA to siRNA. The observed block in dsRNA to siRNA processing in StauC knockdown Lepd-SL1 cells, L. decemlineata tissues and embryonic extracts, and in StauC-deficient Lepd-SL1RR cells supports the role of StauC at the initial stages of RNAi complex formation and processing of dsRNA to siRNA. Further experiments are needed to uncover the precise function of StauC in beetles and to identify substitutes for StauC in other insects. Nevertheless, the discovery of beetle-specific StauC and its conserved role in highly efficient and systemic RNAi response may help to improve RNAi in refractory insects.
The second important finding of this article is that lower levels of StauC in resistant cells compared with susceptible cells are responsible for RNAi resistance observed in Lepd-SL1RR cells. Lepd-SL1 cells were developed from a coleopteran insect, L. decemlineata, pupal tissues (37), and the specific tissue of origin of these cells is not known. The addition of dsRNA to the medium triggers robust RNAi, resulting in an efficient knockdown of the target gene in these cells. The RNAi-resistant Lepd-SL1RR cells were selected by continuous exposure of these cells to dsIAP over multiple generations. In the Lepd-SL1RR cells, dsIAP or other dsRNAs do not induce knockdown of target genes. It is possible that the dsIAP killed the cells that responded to the RNAi treatment, and the remaining cells in the population do not respond to RNAi. Another possibility is that the RNAi-resistant cells selected may have resulted from changes that occurred during exposure to dsRNA over multiple generations. This hypothesis is supported by RNA seq and qRT-PCR data that showed differences in the expression levels of a number of RNAi genes (e.g., StauC, V-ATPase B, and VATPase d) between resistant and susceptible cells. The precise mechanisms used by these cells to develop resistance to RNAi need further investigation.
Dipteran insects including the fruit fly D. melanogaster are refractory to fed or injected dsRNA, but the expression of dsRNA within the cells results in knockdown of target genes (38). Studies in the tephritid fruit fly, Bactrocera dorsalis, showed that transient refractoriness of this insect to fed dsRNA is mediated by changes in the endocytotic pathway (39). Recently, a dsRNA-resistant population of the Western corn rootworm was selected; studies on these insects identified changes to dsRNA uptake as the possible mechanism of resistance (40). Taken together, these studies suggest that dsRNA uptake into cells is one of the potential mechanisms of RNAi resistance. However, changes in uptake of dsRNA into cells do not appear to be a significant contributor to RNAi resistance observed in Lepd-SL1RR cells selected in the current study.
In coleopteran insects, the dsRNA is transported and processed to siRNA efficiently, resulting in robust knockdown of target genes (16). In lepidopteran cells, however, the dsRNA taken up by cells is trapped in the endosomes, resulting in inefficient processing of dsRNA to siRNA, as well as poor knockdown of target genes (16, 17). Recent studies suggest that the trafficking of siRNAs from endosomes into the cytoplasm is a significant hurdle in achieving robust RNAi in gene silencing applications in humans (41). In our experiments, we found that resistant cells take up dsRNA, but it is not processed to siRNA, suggesting that intracellular trafficking or processing of dsRNA to siRNA may have been altered in resistant cells. Collectively, these studies indicate that steps between dsRNA uptake and its processing to siRNA, including intracellular trafficking and processing of dsRNA to siRNA, could be one of the potential mechanisms of RNAi resistance. The fact that StauC is required for the RNAi response and is present in only coleopteran insects, which show robust and systemic RNAi response, suggests that there may be a correlation between the presence of StauC and highly efficient RNAi response. In addition, lower expression of StauC gene was identified as the major factor responsible for RNAi resistance in Lepd-SL1RR cells. Similarly, StauC homologs have not been identified in lepidopteran insects, and their RNAi response is variable and inefficient (11), suggesting a correlation between the presence of StauC and RNAi efficiency. Taken together, these studies contribute to advances in our understanding on the mechanisms of RNAi and provide some insights into potential mechanisms of RNAi resistance.
Materials and Methods
Materials and methods used for cell culture, dsRNA synthesis, gene knockdown, qRT-PCR, RNA sequencing, labeling of dsRNA, phylogenetic analysis, and pull-down and gel shift assays were performed as described in our recent publications (8, 16, 17, 32) and briefly mentioned in the figure legends. Further details on materials and methods used are included in the SI Appendix, and primers used in the experiments are listed in Dataset S3.
Supplementary Material
Acknowledgments
We thank Jeff Howell for help with insect rearing and reading of an earlier version of the manuscript, and Cindy Goodman for sharing Lepd-SL1 cells. Research in the S.R.P. laboratory is supported by grants from the National Institutes of Health (GM070559-12 and 1R21AI131427-01), the National Science Foundation (Industry/University Cooperative Research Centers, the Center for Arthropod Management Technologies under Grant IIP-1338776), and the National Institute of Food and Agriculture, US Department of Agriculture (under HATCH Project 2351177000). This is Publication 18-08-071 from the Kentucky Agricultural Experimental Station and is published with the approval of the director. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. SRP150964).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809381115/-/DCSupplemental.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Gu L, Knipple DC. Recent advances in RNA interference research in insects: Implications for future insect pest management strategies. Crop Prot. 2013;45:36–40. [Google Scholar]
- 3.Scott JG, et al. Towards the elements of successful insect RNAi. J Insect Physiol. 2013;59:1212–1221. doi: 10.1016/j.jinsphys.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Khan SA, Heckel DG, Bock R. Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 2017;35:871–882. doi: 10.1016/j.tibtech.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Bellés X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol. 2010;55:111–128. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- 6.Palli SR. RNAi methods for management of insects and their pathogens. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour. 2012;7:1–10. [Google Scholar]
- 7.Baum JA, et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 8.Zhu F, Xu J, Palli R, Ferguson J, Palli SR. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag Sci. 2011;67:175–182. doi: 10.1002/ps.2048. [DOI] [PubMed] [Google Scholar]
- 9.Palli SR. RNA interference in Colorado potato beetle: Steps toward development of dsRNA as a commercial insecticide. Curr Opin Insect Sci. 2014;6:1–8. doi: 10.1016/j.cois.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomoyasu Y, et al. Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 2008;9:R10. doi: 10.1186/gb-2008-9-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terenius O, et al. RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Singh IK, Singh S, Mogilicherla K, Shukla JN, Palli SR. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci Rep. 2017;7:17059. doi: 10.1038/s41598-017-17134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wynant N, et al. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol. 2014;46:1–8. doi: 10.1016/j.ibmb.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Wynant N, Santos D, Vanden Broeck J. Biological mechanisms determining the success of RNA interference in insects. Int Rev Cell Mol Biol. 2014;312:139–167. doi: 10.1016/B978-0-12-800178-3.00005-1. [DOI] [PubMed] [Google Scholar]
- 15.Guan RB, et al. A nuclease specific to lepidopteran insects suppresses RNAi. J Biol Chem. 2018;293:6011–6021. doi: 10.1074/jbc.RA117.001553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shukla JN, et al. Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 2016;13:656–669. doi: 10.1080/15476286.2016.1191728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon J-S, Gurusamy D, Palli SR. Accumulation of dsRNA in endosomes contributes to inefficient RNA interference in the fall armyworm, Spodoptera frugiperda. Insect Biochem Mol Biol. 2017;90:53–60. doi: 10.1016/j.ibmb.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 19.Lozano J, Gomez-Orte E, Lee HJ, Belles X. Super-induction of Dicer-2 expression by alien double-stranded RNAs: An evolutionary ancient response to viral infection? Dev Genes Evol. 2012;222:229–235. doi: 10.1007/s00427-012-0404-x. [DOI] [PubMed] [Google Scholar]
- 20.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 21.Rubio M, Maestro JL, Piulachs M-D, Belles X. Conserved association of Argonaute 1 and 2 proteins with miRNA and siRNA pathways throughout insect evolution, from cockroaches to flies. Biochim Biophys Acta. 2018;1861:554–560. doi: 10.1016/j.bbagrm.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 23.Schupbach T, Wieschaus E. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol. 1986;113:443–448. doi: 10.1016/0012-1606(86)90179-x. [DOI] [PubMed] [Google Scholar]
- 24.St Johnston D, Beuchle D, Nüsslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 25.Broadus J, Fuerstenberg S, Doe CQ. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- 26.Wickham L, Duchaîne T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3’UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 28.Kiebler MA, et al. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: Implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krichevsky AM, Kosik KS. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 30.St Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alyokhin A, Baker M, Mota-Sanchez D, Dively G, Grafius E. Colorado potato beetle resistance to insecticides. Am J Potato Res. 2008;85:395–413. [Google Scholar]
- 32.Yoon JS, Shukla JN, Gong ZJ, Mogilicherla K, Palli SR. RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: Identification of key contributors. Insect Biochem Mol Biol. 2016;78:78–88. doi: 10.1016/j.ibmb.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Z-M, Tang S, Tao M. Development of resistance to RNAi in mammalian cells. Ann N Y Acad Sci. 2005;1058:105–118. doi: 10.1196/annals.1359.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LeGendre JB, et al. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J Biol Chem. 2013;288:2532–2545. doi: 10.1074/jbc.M112.397349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenna DD. Beetle genomes in the 21st century: Prospects, progress and priorities. Curr Opin Insect Sci. 2018;25:76–82. doi: 10.1016/j.cois.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Long SH, Malntosh A, Grasela J, Goodman CL. The establishment of a Colorado potato beetle (Coleoptera: Chrysomelidae) pupal cell line. Appl Entomol Zool. 2002;37:447–450. [Google Scholar]
- 38.Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Dong X, Zou C, Zhang H. Endocytic pathway mediates refractoriness of insect Bactrocera dorsalis to RNA interference. Sci Rep. 2015;5:8700. doi: 10.1038/srep08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khajuria C, et al. Development and characterization of the first dsRNA-resistant insect population from western corn rootworm, Diabrotica virgifera virgifera LeConte. PLoS One. 2018;13:e0197059. doi: 10.1371/journal.pone.0197059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.