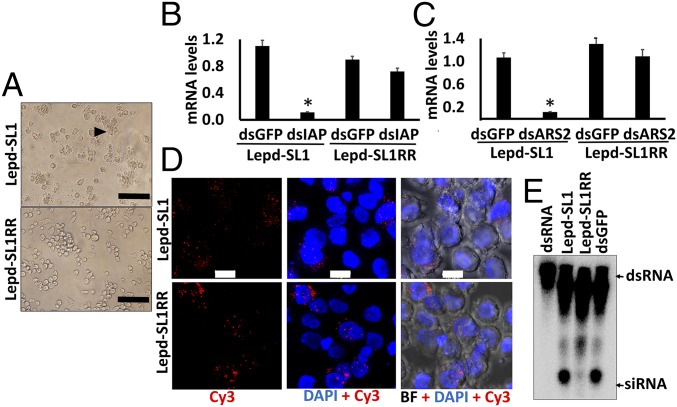
Fig. 4.
Development of the RNAi-resistant Lepd-SL1 cell line and identification of resistance mechanisms. (A) Apoptosis phenotype observed after exposing Lepd-SL1 and Lepd-SL1RR cells to dsIAP. The cells were exposed to 20 ng dsIAP in 100 µL medium and photographed at 24 h after treatment. The arrow points to the cells showing apoptosis phenotype. (Scale bar, 100 μm.) (B) Relative IAP mRNA levels in Lepd-SL1 and Lepd-SL1RR cells exposed to dsIAP. The cells were exposed to dsIAP or dsGFP (a control), and total RNA was isolated and used to quantify relative IAP mRNA levels by qRT-PCR. Ribosomal protein 4 (RP4) was used as an internal control. The bars show mean ± SD (n = 3). *Significantly different from control at P ≤ 0.05. (C) Relative Ars2 mRNA levels in Lepd-SL1 and Lepd-SL1RR cells exposed to dsArs2 quantified as described in B legend. (D) Subcellular localization of Cy3-labeled dsGFP in the Lepd-SL1 and Lepd-SL1RR cells. The cells were exposed to 25 ng Cy3-labeled dsGFP in 8-well chamber slides for 2 h. Then the cells were fixed, mounted in DAPI containing medium and photographed using a confocal microscope. BF, bright field; Cy3, Cy3-labeled dsGFP. (Scale bar, 10 μm.) (E) Comparison of processing of dsRNA to siRNA in Lepd-SL1 and Lepd-SL1RR cells. The cells in six-well plates were exposed to 1.6 million cpm 32P-labeled dsGFP. At 24 h after the addition of dsRNA, the cells were harvested and RNA was isolated. RNA containing 2,000 cpm was resolved on 16% acrylamide-urea gel. The first and last lanes show GFP dsRNA and GFP dsRNA digested to siRNA with RNase III, respectively, used as markers. The arrows point to dsRNA and siRNA bands.